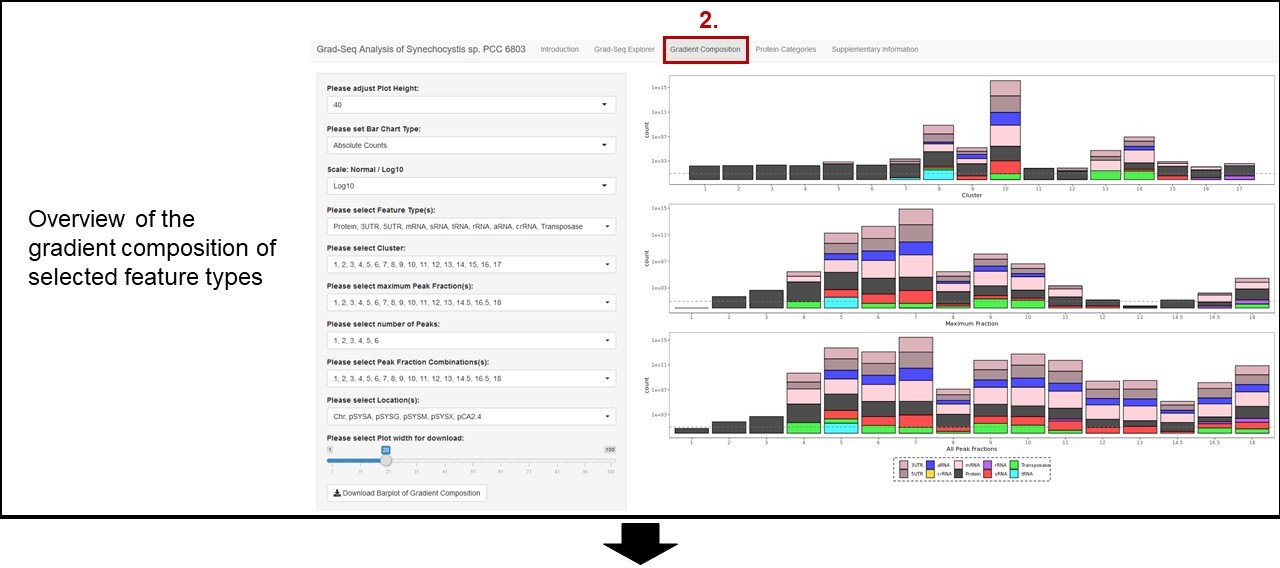
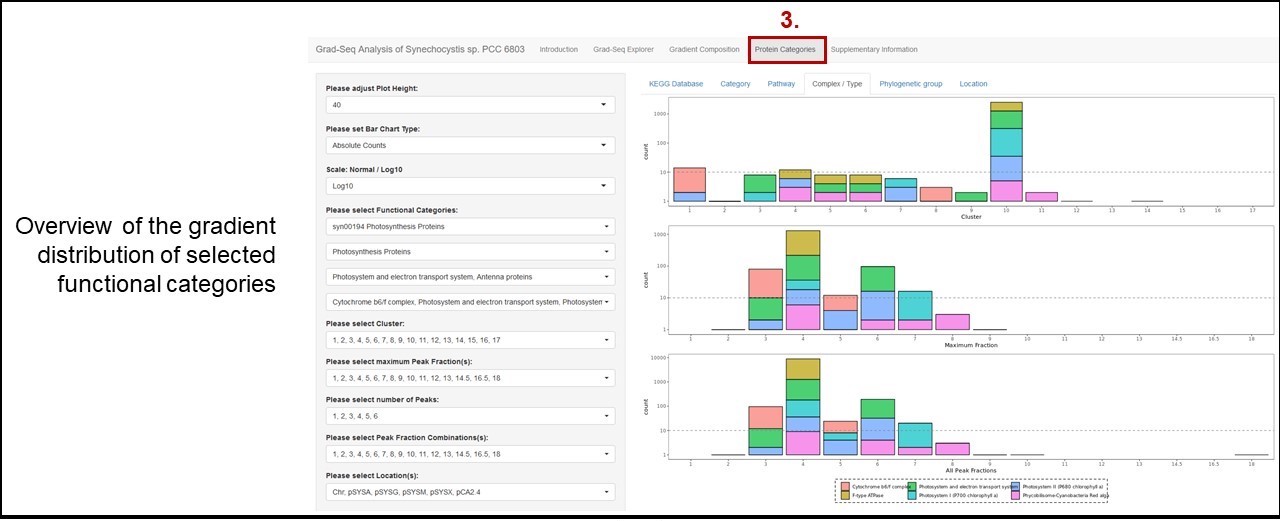
Please choose Gene Identifier:
16S rRNA/6803r03 (rRNA)
16S rRNA/6803r04 (rRNA)
23S rRNA/6803r02 (rRNA)
23S rRNA/6803r05 (rRNA)
5S rRNA/6803r01 (rRNA)
5S rRNA/6803r06 (rRNA)
TU241 (6803t02, ncl0080, sll1670) - Transcr Unit
TU256 (6803t03) - Transcr Unit
TU317 (6803t04) - Transcr Unit
TU430 (6803t06, slr0695, slr1590) - Transcr Unit
TU629 (6803t07) - Transcr Unit
TU641 (6803t08, sll0990, sll0992) - Transcr Unit
TU1004 (6803t10) - Transcr Unit
TU1111 (6803t11) - Transcr Unit
TU1587 (6803t15) - Transcr Unit
TU1663 (6803t16, ncr0650) - Transcr Unit
TU1780 (6803t18) - Transcr Unit
TU1890 (6803t19) - Transcr Unit
TU1904 (6803t20) - Transcr Unit
TU1910 (6803t21) - Transcr Unit
TU2132 (6803t22) - Transcr Unit
TU2175 (6803t24) - Transcr Unit
TU2230 (6803t25) - Transcr Unit
TU2321 (6803t26) - Transcr Unit
TU2505 (6803t28) - Transcr Unit
TU2613 (6803t31) - Transcr Unit
TU2719 (6803t32) - Transcr Unit
TU2792 (6803t33) - Transcr Unit
TU2935 (6803t34, slr0915, slr0917) - Transcr Unit
TU3394 (6803t37, 6803t38) - Transcr Unit
TU3465 (6803t39) - Transcr Unit
TU3514 (6803t41) - Transcr Unit
TU1192 (sll0869) - Transcr Unit
accA/sll0728 (mRNA / CDS) - TU3600
accA/sll0728 (Protein) - TU3600
TU3600 (sll0728, sll0729) - Transcr Unit
accB/slr0435 (mRNA / CDS) - TU2674
accB/slr0435 (Protein) - TU2674
accC/sll0053 (mRNA / CDS) - TU2735
accC/sll0053 (Protein) - TU2735
TU2735 (sll0053) - Transcr Unit
accD, ycf11, zfpA/sll0336 (mRNA / CDS) - TU2461
accD, ycf11, zfpA/sll0336 (Protein) - TU2461
TU2461 (sll0336) - Transcr Unit
ackA/sll1299 (mRNA / CDS) - TU673
ackA/sll1299 (Protein) - TU673
acnB/slr0665 (mRNA / CDS) - TU3530
acnB/slr0665 (Protein) - TU3530
acp/ssl2084 (mRNA / CDS) - TU823
acpP/ssl2084 (Protein) - TU823
acs/sll0542 (mRNA / CDS) - TU2968
acs/sll0542 (Protein) - TU2968
sll0542-3UTR (3'UTR) - TU2968
act/sll1900 (mRNA / CDS) - TU1817
act/sll1900 (Protein) - TU1817
adk/sll1059 (mRNA / CDS) - TU72
adk/sll1059 (Protein) - TU72
adk/sll1815 (mRNA / CDS) - TU836
adk/sll1815 (Protein) - TU836
ahcY/sll1234 (mRNA / CDS) - TU1699
ahcY/sll1234 (Protein) - TU1699
sll1234-3UTR (3'UTR) - TU1699
alaS/sll0362 (mRNA / CDS) - TU2220
alaS/sll0362 (Protein) - TU2220
TU2220 (sll0362) - Transcr Unit
sll0362-3UTR (3'UTR) - TU2220
ama/sll0100 (mRNA / CDS) - TU3150
ama/sll0100 (Protein) - TU3150
ama/slr1653 (mRNA / CDS) - TU3629
ama/slr1653 (Protein) - TU3629
amiA/slr0891 (mRNA / CDS)
amiA/slr0891 (Protein)
amiA/slr1744 (mRNA / CDS) - TU1369
amiA/slr1744 (Protein) - TU1369
slr1744-5UTR (5'UTR) - TU1369
slr1744-as (asRNA)
ams1/slr0323 (mRNA / CDS) - TU2368
ams1/slr0323 (Protein) - TU2368
amt1/sll0108 (mRNA / CDS) - TU3138
amt1/sll0108 (Protein) - TU3138
amt2/sll1017 (mRNA / CDS) - TU384
amt2/sll1017 (Protein) - TU384
TU384 (sll1017, sll1018) - Transcr Unit
Ank/slr1109 (mRNA / CDS) - TU212
Ank/slr1109 (Protein) - TU212
anmK/slr1179 (mRNA / CDS) - TU3680
anmK/slr1179 (Protein) - TU3680
apcA/slr2067 (mRNA / CDS) - TU1472
apcC/ssr3383 (Protein) - TU1472
slr2067-5UTR (5'UTR) - TU1472
TU1472 (slr2067, slr1986, ssr3383, SyR2) - Transcr Unit
apcB/slr1986 (mRNA / CDS) - TU1472
cpcD/ssl3093 (Protein) - TU716
apcC/ssr3383 (mRNA / CDS) - TU1472
apcA/slr2067 (Protein) - TU1472
apcD/sll0928 (mRNA / CDS) - TU333
apcB/slr1986 (Protein) - TU1472
TU333 (sll0928, sll0930, sll0931, sll0932) - Transcr Unit
apcE/slr0335 (mRNA / CDS) - TU2385
apcD/sll0928 (Protein) - TU333
TU2384 (slr0335) - Transcr Unit
TU2385 (slr0335) - Transcr Unit
slr0335-3UTR (3'UTR) - TU2385
slr0335-5UTR (5'UTR) - TU2385
apcF/slr1459 (mRNA / CDS) - TU3482
cpcA/sll1578 (Protein) - TU716
TU3482 (slr1459) - Transcr Unit
slr1459-3UTR (3'UTR) - TU3482
APE2/sll1343 (mRNA / CDS) - TU3442
APE2/sll1343 (Protein) - TU3442
apt/sll1430 (Protein) - TU1613
argB/slr1898 (mRNA / CDS) - TU857
argB/slr1898 (Protein) - TU857
TU857 (slr1898) - Transcr Unit
argC/sll0080 (mRNA / CDS) - TU3047
argC/sll0080 (Protein) - TU3047
TU3047 (sll0080) - Transcr Unit
sll0080-3UTR (3'UTR) - TU3047
argD/slr1022 (mRNA / CDS) - TU638
argD/slr1022 (Protein) - TU638
TU638 (slr1022) - Transcr Unit
slr1022-3UTR (3'UTR) - TU638
slr1022-5UTR (5'UTR) - TU638
argF/sll0902 (mRNA / CDS) - TU3396
argF/sll0902 (Protein) - TU3396
TU3396 (sll0902, sll0493, sll0494) - Transcr Unit
argG/slr0585 (mRNA / CDS) - TU3688
argG/slr0585 (Protein) - TU3688
TU3688 (slr0585) - Transcr Unit
argH/slr1133 (mRNA / CDS) - TU786
argH/slr1133 (Protein) - TU786
argJ/sll1883 (mRNA / CDS) - TU1860
argJ/sll1883 (Protein) - TU1860
TU1860 (sll1883) - Transcr Unit
sll1883-5UTR (5'UTR) - TU1860
argS/sll0502 (mRNA / CDS) - TU3381
argS/sll0502 (Protein) - TU3381
sll0502-5UTR (5'UTR) - TU3381
TU3381 (sll0502, sll0503) - Transcr Unit
aroA/slr0444 (mRNA / CDS) - TU2171
aroA/slr0444 (Protein) - TU2171
TU2171 (slr0444) - Transcr Unit
TU2172 (slr0444) - Transcr Unit
aroB/slr2130 (mRNA / CDS) - TU1682
aroB/slr2130 (Protein) - TU1682
TU1682 (slr2130) - Transcr Unit
aroC/sll1747 (mRNA / CDS) - TU930
aroC/sll1747 (Protein) - TU930
TU930 (sll1747, ssl3712) - Transcr Unit
aroE/slr1559 (mRNA / CDS) - TU3547
aroE/slr1559 (Protein) - TU3547
TU3547 (slr1559) - Transcr Unit
aroH/sll0109 (mRNA / CDS) - TU3134
aroH/sll0109 (Protein) - TU3134
TU3134 (sll0109) - Transcr Unit
sll0109-5UTR (5'UTR) - TU3134
aroK/sll1669 (mRNA / CDS) - TU243
aroK/sll1669 (Protein) - TU243
TU243 (sll1669) - Transcr Unit
aroQ/sll1112 (mRNA / CDS) - TU908
aroQ/sll1112 (Protein) - TU908
arsA/sll1957 (Protein) - TU1452
arsC/slr0946 (Protein)
TU158 (As1_flv4) - Transcr Unit
asd/slr0549 (mRNA / CDS) - TU3417
asd/slr0549 (Protein) - TU3417
aslB/sll1766 (mRNA / CDS) - TU1262
aslB/sll1766 (Protein) - TU1262
asnS/sll0495 (mRNA / CDS)
asnS/sll0495 (Protein)
aspC/sll0006 (mRNA / CDS) - TU2604
aspC/sll0006 (Protein) - TU2604
sll0006-as (asRNA)
aspC/sll0402 (mRNA / CDS) - TU2688
aspC/sll0402 (Protein) - TU2688
TU2688 (sll0402) - Transcr Unit
sll0402-5UTR (5'UTR) - TU2688
aspS/slr1720 (mRNA / CDS) - TU1506
aspS/slr1720 (Protein) - TU1506
AT103/sll1874 (mRNA / CDS)
AT103/sll1874 (Protein)
atpA/sll1326 (mRNA / CDS) - TU166
atpA/sll1326 (Protein) - TU166
atpB/slr1329 (mRNA / CDS) - TU1709
atpB/slr1329 (Protein) - TU1709
TU1709 (slr1329) - Transcr Unit
slr1329-3UTR (3'UTR) - TU1709
atpC/sll1327 (mRNA / CDS) - TU166
atpC/sll1327 (Protein) - TU166
atpD/sll1325 (mRNA / CDS) - TU166
atpD/sll1325 (Protein) - TU166
atpE/slr1330 (mRNA / CDS) - TU1711
atpE/slr1330 (Protein) - TU1711
slr1330-5UTR (5'UTR) - TU1711
TU1711 (slr1330, slr1331, slr1332) - Transcr Unit
atpF/sll1324 (mRNA / CDS) - TU166
atpF/sll1324 (Protein) - TU166
atpG/sll1323 (mRNA / CDS) - TU166
atpG/sll1323 (Protein) - TU166
atpH/ssl2615 (mRNA / CDS) - TU166
atpH/ssl2615 (Protein) - TU166
atpI/sll1322 (mRNA / CDS) - TU166
atpI/sll1322 (Protein) - TU166
atx1/ssr2857 (mRNA / CDS) - TU737
atx1/ssr2857 (Protein) - TU737
TU737 (ssr2857) - Transcr Unit
ssr2857-5UTR (5'UTR) - TU737
bchB/ssr2049 (Protein) - TU1067
bcp/sll0221 (mRNA / CDS) - TU152
bcp/sll0221 (Protein) - TU152
bcp/slr0242 (mRNA / CDS) - TU127
bcp/slr0242 (Protein) - TU127
bfr/sll1341 (mRNA / CDS) - TU3450
bfr/sll1341 (Protein) - TU3450
sll1341-3UTR (3'UTR) - TU3450
sll1341-5UTR (5'UTR) - TU3450
bfr/slr1890 (mRNA / CDS)
bfr/slr1890 (Protein)
bgl/sll1538 (mRNA / CDS) - TU2117
bgl/sll1538 (Protein) - TU2117
bgtA/slr1735 (mRNA / CDS) - TU1350
bgtA/slr1735 (Protein) - TU1350
TU1350 (slr1735) - Transcr Unit
slr1735-5UTR (5'UTR) - TU1350
bgtB/sll1270 (mRNA / CDS) - TU1141
bgtB/sll1270 (Protein) - TU1141
sll1270-5UTR (5'UTR) - TU1141
TU1141 (sll1270, sll1271, ssl2507, sll1272) - Transcr Unit
bicA/sll0834 (mRNA / CDS) - TU2989
bicA/sll0834 (Protein) - TU2989
sll0834-3UTR (3'UTR) - TU2989
bioB/slr1364 (mRNA / CDS) - TU1139
bioB/slr1364 (Protein) - TU1139
TU1139 (slr1364, slr1365, slr1366) - Transcr Unit
bioD/slr0523 (mRNA / CDS) - TU3103
bioD/slr0523 (Protein) - TU3103
slr0523-5UTR (5'UTR) - TU3103
bioF/slr0917 (mRNA / CDS) - TU2936
bioF/slr0917 (Protein) - TU2936
slr0917-3UTR (3'UTR) - TU2935
birA/sll1655 (mRNA / CDS) - TU270
birA/sll1655 (Protein) - TU270
btpA/sll0634 (mRNA / CDS) - TU429
btpA/sll0634 (Protein) - TU429
TU429 (sll0634, sll0635) - Transcr Unit
bvdR/slr1784 (mRNA / CDS) - TU229
bvdR/slr1784 (Protein) - TU229
TU229 (slr1784) - Transcr Unit
slr1784-5UTR (5'UTR) - TU229
cad/sll1683 (mRNA / CDS)
cad/sll1683 (Protein)
capA/slr1764 (mRNA / CDS)
capA/slr1764 (Protein)
CARA/sll1498 (mRNA / CDS) - TU3538
CARA/sll1498 (Protein) - TU3538
cbaB/sll1869 (mRNA / CDS) - TU1876
cbaB/sll1869 (Protein) - TU1876
cbiX/sll0037 (mRNA / CDS) - TU3320
cbiX/sll0037 (Protein) - TU3320
sll0037-5UTR (5'UTR) - TU3320
TU3528 (sll0621, sll0622) - Transcr Unit
ccmA/sll0934 (mRNA / CDS) - TU331
ccmA/sll0934 (Protein) - TU331
ccmK1/sll1029 (mRNA / CDS) - TU205
ccmK1/sll1029 (Protein) - TU205
ccmK2/sll1028 (mRNA / CDS) - TU205
ccmK2/sll1028 (Protein) - TU205
sll1028-as (asRNA)
ccmK3/slr1838 (mRNA / CDS) - TU968
ccmK3/slr1838 (Protein) - TU968
TU968 (slr1838, slr1839, slr1840) - Transcr Unit
ccmK4/slr1839 (mRNA / CDS) - TU968
ccmK4/slr1839 (Protein) - TU968
ccmL/sll1030 (mRNA / CDS) - TU205
ccmL/sll1030 (Protein) - TU205
ccmM/sll1031 (mRNA / CDS) - TU205
ccmM/sll1031 (Protein) - TU205
ccmN/sll1032 (mRNA / CDS) - TU205
ccmN/sll1032 (Protein) - TU205
ccmO/slr0436 (mRNA / CDS) - TU2674
ccmO/slr0436 (Protein) - TU2674
ccs1, ycf44/slr2087 (mRNA / CDS) - TU1583
ccs1, ycf44/slr2087 (Protein) - TU1583
TU1583 (slr2087) - Transcr Unit
ccsA, ycf5/sll1513 (mRNA / CDS) - TU446
ccsA, ycf5/sll1513 (Protein) - TU446
sll1513-3UTR (3'UTR) - TU446
sll1513-5UTR (5'UTR) - TU446
TU446 (sll1513) - Transcr Unit
TU1152 (slr1369) - Transcr Unit
cefD/slr2143 (mRNA / CDS) - TU1306
cefD/slr2143 (Protein) - TU1306
slr2143-3UTR (3'UTR) - TU1306
cheW/slr1043 (mRNA / CDS) - TU481
cheW/slr1043 (Protein) - TU481
chlB/slr0772 (mRNA / CDS) - TU2490
chlB/slr0772 (Protein) - TU2490
TU2490 (slr0772) - Transcr Unit
chlD/slr1777 (mRNA / CDS) - TU221
chlD/slr1777 (Protein) - TU221
slr1777-5UTR (5'UTR) - TU221
TU221 (slr1777, ssr2975, slr1778) - Transcr Unit
TU220 (slr1777, ssr2975, slr1778) - Transcr Unit
chlG/slr0056 (mRNA / CDS) - TU2713
chlG/slr0056 (Protein) - TU2713
TU2713 (slr0056) - Transcr Unit
chlH/slr1055 (mRNA / CDS) - TU496
chlH/slr1055 (Protein) - TU496
TU496 (slr1055) - Transcr Unit
slr1055-3UTR (3'UTR) - TU496
chlI/slr1030 (mRNA / CDS) - TU656
chlI/slr1030 (Protein) - TU656
TU656 (slr1030, slr1031, slr1032, slr1033, ssr1720) - Transcr Unit
TU3594 (slr0749, ssr1251, slr0750) - Transcr Unit
chlM/slr0525 (mRNA / CDS) - TU3346
chlM/slr0525 (Protein) - TU3346
TU3346 (slr0525, slr0526, slr0527) - Transcr Unit
chlN/slr0750 (mRNA / CDS) - TU3594
chlN/slr0750 (Protein) - TU3594
chlP/sll1091 (mRNA / CDS) - TU2008
chlP/sll1091 (Protein) - TU2008
TU2008 (sll1091) - Transcr Unit
sll1091-3UTR (3'UTR) - TU2008
citH/sll0891 (mRNA / CDS) - TU2940
citH/sll0891 (Protein) - TU2940
clcD/sll1298 (mRNA / CDS) - TU673
clcD/sll1298 (Protein) - TU673
clpB1/slr1641 (mRNA / CDS) - TU2138
clpB1/slr1641 (Protein) - TU2138
TU2138 (slr1641) - Transcr Unit
slr1641-3UTR (3'UTR) - TU2138
clpB2/slr0156 (mRNA / CDS) - TU2280
clpB2/slr0156 (Protein) - TU2280
TU2280 (slr0156, slr0157) - Transcr Unit
clpC/sll0020 (mRNA / CDS) - TU2570
clpC/sll0020 (Protein) - TU2570
sll0020-3UTR (3'UTR) - TU2570
sll0020-5UTR (5'UTR) - TU2570
clpP/slr0542 (mRNA / CDS) - TU3385
clpP/slr0542 (Protein) - TU3385
TU3385 (slr0542, slr0543) - Transcr Unit
clpP2/sll0534 (mRNA / CDS) - TU3414
clpP2/sll0534 (Protein) - TU3414
clpP3/slr0165 (mRNA / CDS) - TU2285
clpP3/slr0165 (Protein) - TU2285
clpP4/slr0164 (mRNA / CDS) - TU2285
clpP4/slr0164 (Protein) - TU2285
slr0164-5UTR (5'UTR) - TU2285
TU2284 (slr0164, slr0165) - Transcr Unit
TU2285 (slr0164, slr0165) - Transcr Unit
clpS/ssl3379 (mRNA / CDS) - TU1256
clpS/ssl3379 (Protein) - TU1256
ssl3379-3UTR (3'UTR) - TU1256
clpS/ssr2723 (mRNA / CDS) - TU2109
clpS/ssr2723 (Protein) - TU2109
ssr2723-3UTR (3'UTR) - TU2109
clpX/sll0535 (mRNA / CDS) - TU3414
clpX/sll0535 (Protein) - TU3414
cmpA/slr0040 (mRNA / CDS) - TU3332
cmpA/slr0040 (Protein) - TU3332
slr0040-5UTR (5'UTR) - TU3332
TU3332 (slr0040, slr0041, slr0042, slr0043, slr0044) - Transcr Unit
cmpB/slr0041 (mRNA / CDS) - TU3332
cmpB/slr0041 (Protein) - TU3332
cmpC/slr0043 (mRNA / CDS) - TU3332
cmpC/slr0043 (Protein) - TU3332
cmpD/slr0044 (mRNA / CDS) - TU3332
cmpD/slr0044 (Protein) - TU3332
cmpR/sll0030 (mRNA / CDS) - TU3331
cmpR/sll0030 (Protein) - TU3331
TU3331 (sll0030) - Transcr Unit
sll0030-5UTR (5'UTR) - TU3331
cnfU, synifU/ssl2667 (mRNA / CDS) - TU3497
cnfU, synifU/ssl2667 (Protein) - TU3497
ssl2667-5UTR (5'UTR) - TU3497
TU3497 (ssl2667, ssrA/tmRNA) - Transcr Unit
coaE/slr0553 (mRNA / CDS) - TU3419
coaE/slr0553 (Protein) - TU3419
coaX/slr0812 (Protein) - TU1744
TU2852 (sll0378) - Transcr Unit
cobA/hemD/sll0166 (mRNA / CDS) - TU2410
cobA/hemD/sll0166 (Protein) - TU2410
cobH/sll0916 (Protein)
cobI/slr1879 (Protein)
cobJ/slr0969 (mRNA / CDS) - TU328
cobJ/slr0969 (Protein) - TU328
TU328 (slr0969) - Transcr Unit
cobL/slr1368 (mRNA / CDS) - TU1151
cobL/slr1368 (Protein) - TU1151
TU1151 (slr1368) - Transcr Unit
cbiF/slr0239 (mRNA / CDS) - TU124
cobM/slr0239 (Protein) - TU124
TU124 (slr0239) - Transcr Unit
slr0239-5UTR (5'UTR) - TU124
cobN/slr1211 (mRNA / CDS)
cobN/slr1211 (Protein)
cobA/slr0260 (mRNA / CDS) - TU1541
cobO/slr0260 (Protein) - TU1541
TU1541 (slr0260) - Transcr Unit
cobU/slr0216 (mRNA / CDS) - TU2618
cobP/slr0216 (Protein) - TU2618
TU2620 (slr0216) - Transcr Unit
cobQ/slr0618 (mRNA / CDS) - TU3117
cobQ/slr0618 (Protein) - TU3117
TU3117 (slr0618) - Transcr Unit
cobW/slr0502 (mRNA / CDS) - TU3071
cobW/slr0502 (Protein) - TU3071
codA/slr1237 (mRNA / CDS) - TU1077
codA/slr1237 (Protein) - TU1077
slr1237-as (asRNA)
cofH/sll1659 (mRNA / CDS) - TU261
cofH/sll1659 (Protein) - TU261
comA/slr0197 (mRNA / CDS) - TU2881
comA/slr0197 (Protein) - TU2881
slr0197-as (asRNA)
comB/slr1718 (mRNA / CDS) - TU1505
comB/slr1718 (Protein) - TU1505
TU595 (sll1929) - Transcr Unit
comM/slr0904 (mRNA / CDS) - TU2927
comM/slr0904 (Protein) - TU2927
slr0904-5UTR (5'UTR) - TU2927
TU2927 (slr0904, slr0905) - Transcr Unit
corA/sll0507 (mRNA / CDS) - TU3374
corA/sll0507 (Protein) - TU3374
corA/sll0671 (mRNA / CDS)
corA/sll0671 (Protein)
TU3221 (sll0794) - Transcr Unit
cp12/ssl3364 (mRNA / CDS) - TU1273
cp12/ssl3364 (Protein) - TU1273
TU1273 (ssl3364) - Transcr Unit
ssl3364-3UTR (3'UTR) - TU1273
ssl3364-5UTR (5'UTR) - TU1273
cpcA/sll1578 (mRNA / CDS) - TU716
apcF/slr1459 (Protein) - TU3482
cpcB/sll1577 (mRNA / CDS) - TU716
cpcB/sll1577 (Protein) - TU716
sll1577-5UTR (5'UTR) - TU716
TU716 (sll1577, sll1578, sll1579, sll1580, ssl3093) - Transcr Unit
TU718 (sll1577, sll1578, sll1579, sll1580, ssl3093) - Transcr Unit
TU719 (sll1577, sll1578, sll1579, sll1580, ssl3093) - Transcr Unit
cpcC1/sll1580 (mRNA / CDS) - TU716
cpcS1/sll0853 (Protein) - TU1381
cpcC2/sll1579 (mRNA / CDS) - TU716
cpcT/slr1649 (Protein) - TU2153
cpcD/ssl3093 (mRNA / CDS) - TU716
cpcF/sll1051 (Protein) - TU82
cpcE/slr1878 (mRNA / CDS) - TU1260
cpcG1/slr2051 (Protein) - TU584
cpcF/sll1051 (mRNA / CDS) - TU82
cpcG2/sll1471 (Protein) - TU3579
TU82 (sll1051, sll1052) - Transcr Unit
cpcG1/slr2051 (mRNA / CDS) - TU584
cpcE/slr1878 (Protein) - TU1260
TU584 (slr2051) - Transcr Unit
slr2051-3UTR (3'UTR) - TU584
slr2051-5UTR (5'UTR) - TU584
cpcG2/sll1471 (mRNA / CDS) - TU3579
cpcC2/sll1579 (Protein) - TU716
TU3579 (sll1471, sll1472, sll1473, sll1474, sll1475) - Transcr Unit
cpcS1/sll0853 (mRNA / CDS) - TU1381
cpcC1/sll1580 (Protein) - TU716
cpcT/slr1649 (mRNA / CDS) - TU2153
napC/sll0873 (Protein)
cph1, hik35/slr0473 (mRNA / CDS) - TU2746
cph1, hik35/slr0473 (Protein) - TU2746
cph2/sll0821 (mRNA / CDS) - TU3022
cph2/sll0821 (Protein) - TU3022
cphA/slr2002 (mRNA / CDS) - TU1488
cphA/slr2002 (Protein) - TU1488
slr2002-5UTR (5'UTR) - TU1488
TU1488 (slr2002, slr2003) - Transcr Unit
cphB/slr2001 (mRNA / CDS) - TU1485
cphB/slr2001 (Protein) - TU1485
TU1485 (slr2001) - Transcr Unit
slr2001-3UTR (3'UTR) - TU1485
cpmA/sll1489 (mRNA / CDS) - TU3559
cpmA/sll1489 (Protein) - TU3559
cpx, katG/sll1987 (mRNA / CDS) - TU1578
cpx, katG/sll1987 (Protein) - TU1578
TU1578 (sll1987) - Transcr Unit
crhR/slr0083 (mRNA / CDS) - TU3046
crhR/slr0083 (Protein) - TU3046
TU3046 (slr0083) - Transcr Unit
slr0083-3UTR (3'UTR) - TU3046
slr0083-5UTR (5'UTR) - TU3046
crtE/slr0739 (mRNA / CDS) - TU114
crtE/slr0739 (Protein) - TU114
TU114 (slr0739) - Transcr Unit
crtH/sll0033 (mRNA / CDS) - TU3326
crtH/sll0033 (Protein) - TU3326
TU3326 (sll0033) - Transcr Unit
crtO/slr0088 (mRNA / CDS) - TU3054
crtO/slr0088 (Protein) - TU3054
crtQ-2/slr0940 (mRNA / CDS) - TU2078
crtQ-2/slr0940 (Protein) - TU2078
TU2078 (slr0940, slr0941) - Transcr Unit
TU998 (sll1468) - Transcr Unit
crtX/slr1125 (mRNA / CDS) - TU80
crtX/slr1125 (Protein) - TU80
cscK/slr1448 (mRNA / CDS) - TU1118
cscK/slr1448 (Protein) - TU1118
csgA/sll1704 (Protein)
TU905 (csiR1, slr1214) - Transcr Unit
TU2348 (slr1950, slr1951) - Transcr Unit
ctaC/sll0813 (mRNA / CDS) - TU1742
ctaC/sll0813 (Protein) - TU1742
TU1742 (sll0813) - Transcr Unit
ctaCI/slr1136 (mRNA / CDS) - TU791
ctaCI/slr1136 (Protein) - TU791
TU791 (slr1136, slr1137, slr1138) - Transcr Unit
ctaDI/slr1137 (mRNA / CDS) - TU791
ctaDI/slr1137 (Protein) - TU791
ctaDII/slr2082 (mRNA / CDS) - TU1573
ctaDII/slr2082 (Protein) - TU1573
TU1573 (slr2082, slr2083, slr2084) - Transcr Unit
ctpA/slr0008 (mRNA / CDS) - TU2580
ctpA/slr0008 (Protein) - TU2580
TU2580 (slr0008) - Transcr Unit
slr0008-5UTR (5'UTR) - TU2580
ctpB/slr0257 (mRNA / CDS) - TU162
ctpB/slr0257 (Protein) - TU162
ctr1/slr1044 (mRNA / CDS) - TU481
ctr1/slr1044 (Protein) - TU481
cupA/sll1734 (mRNA / CDS) - TU951
cupA/sll1734 (Protein) - TU951
cupB/slr1302 (mRNA / CDS) - TU303
cupB/slr1302 (Protein) - TU303
TU303 (slr1302, slr1303) - Transcr Unit
cya1/slr1991 (mRNA / CDS) - TU1476
cya1/slr1991 (Protein) - TU1476
cya2/sll0646 (mRNA / CDS) - TU406
cya2/sll0646 (Protein) - TU406
cydA/slr1379 (mRNA / CDS) - TU672
cydA/slr1379 (Protein) - TU672
cynS/slr0899 (mRNA / CDS) - TU2925
cynS/slr0899 (Protein) - TU2925
cyp/slr0574 (mRNA / CDS) - TU2965
cyp/slr0574 (Protein) - TU2965
cyp/slr1251 (mRNA / CDS) - TU1430
cyp/slr1251 (Protein) - TU1430
sll1041 (mRNA / CDS) - TU178
cysA/sll1041 (Protein) - TU178
cysC/slr0676 (mRNA / CDS) - TU393
cysC/slr0676 (Protein) - TU393
TU393 (slr0676, slr0677, slr0678, slr0679) - Transcr Unit
cysE/slr1348 (mRNA / CDS) - TU1800
cysE/slr1348 (Protein) - TU1800
cysH/slr1791 (Protein) - TU238
cysK/slr1842 (mRNA / CDS) - TU972
cysK/slr1842 (Protein) - TU972
TU972 (slr1842) - Transcr Unit
cysM/sll0712 (mRNA / CDS) - TU96
cysM/sll0712 (Protein) - TU96
TU96 (sll0712) - Transcr Unit
cysQ/sll0895 (mRNA / CDS) - TU3410
cysQ/sll0895 (Protein) - TU3410
cysS/slr0958 (mRNA / CDS) - TU2102
cysS/slr0958 (Protein) - TU2102
TU2102 (slr0958) - Transcr Unit
TU1807 (sll1245) - Transcr Unit
dacB/slr0646 (mRNA / CDS) - TU2821
dacB/slr0646 (Protein) - TU2821
dacB/slr0804 (mRNA / CDS) - TU1725
dacB/slr0804 (Protein) - TU1725
dapA/slr0550 (mRNA / CDS) - TU3417
dapA/slr0550 (Protein) - TU3417
dapB/sll1058 (mRNA / CDS) - TU72
dapB/sll1058 (Protein) - TU72
TU72 (sll1058, sll1059) - Transcr Unit
dapF/slr1665 (mRNA / CDS) - TU3649
dapF/slr1665 (Protein) - TU3649
TU3649 (slr1665) - Transcr Unit
slr1665-5UTR (5'UTR) - TU3649
dapL/sll0480 (mRNA / CDS) - TU3077
dapL/sll0480 (Protein) - TU3077
dcd/sll1258 (Protein) - TU1782
dctP/sll1314 (mRNA / CDS)
dctP/sll1314 (Protein)
ddh/slr1556 (mRNA / CDS) - TU3535
ddh/slr1556 (Protein) - TU3535
slr1556-5UTR (5'UTR) - TU3535
slr1556-as (asRNA)
ddl/slr1874 (mRNA / CDS) - TU1254
ddl/slr1874 (Protein) - TU1254
ddpX/slr1679 (Protein) - TU2028
def/slr1549 (mRNA / CDS) - TU1014
def/slr1549 (Protein) - TU1014
slr1549-5UTR (5'UTR) - TU1014
degT/slr1666 (mRNA / CDS) - TU3651
degT/slr1666 (Protein) - TU3651
deoC/sll1776 (mRNA / CDS) - TU1237
deoC/sll1776 (Protein) - TU1237
TU1803 (slr1350) - Transcr Unit
TU1965 (sll1441) - Transcr Unit
desC,des9/sll0541 (mRNA / CDS) - TU2970
desC,des9/sll0541 (Protein) - TU2970
sll0541-3UTR (3'UTR) - TU2970
sll0541-5UTR (5'UTR) - TU2970
TU2970 (sll0541) - Transcr Unit
TU2196 (sll0262) - Transcr Unit
devB/sll1479 (mRNA / CDS) - TU3574
devB/sll1479 (Protein) - TU3574
sll1479-3UTR (3'UTR) - TU3574
dfa1/sll0550 (mRNA / CDS) - TU2954
dfa1/sll0550 (Protein) - TU2954
sll0550-3UTR (3'UTR) - TU2954
dfa2/sll0217 (mRNA / CDS) - TU156
dfa2/sll0217 (Protein) - TU156
sll0217-as2 (asRNA)
dfa3/sll1521 (mRNA / CDS)
dfa3/sll1521 (Protein)
dfa4/sll0219 (mRNA / CDS) - TU156
dfa4/sll0219 (Protein) - TU156
dfp/sll0250 (mRNA / CDS) - TU1554
dfp/sll0250 (Protein) - TU1554
dfrA/slr1706 (mRNA / CDS) - TU736
dfrA/slr1706 (Protein) - TU736
dgt/sll0398 (mRNA / CDS) - TU2831
dgt/sll0398 (Protein) - TU2831
divK/slr2041 (mRNA / CDS) - TU561
divK/slr2041 (Protein) - TU561
dnaB/slr0833 (mRNA / CDS) - TU2993
dnaB/slr0833 (Protein) - TU2993
dnaE/slr0603 (mRNA / CDS) - TU3709
dnaE/slr0603 (Protein) - TU3709
TU3709 (slr0603) - Transcr Unit
dnaJ/sll0897 (mRNA / CDS) - TU3409
dnaJ/sll0897 (Protein) - TU3409
dnaJ/sll1666 (mRNA / CDS) - TU248
dnaJ/sll1666 (Protein) - TU248
dnaJ/sll1933 (mRNA / CDS) - TU588
dnaJ/sll1933 (Protein) - TU588
dnaJ/slr0093 (mRNA / CDS) - TU3059
dnaJ/slr0093 (Protein) - TU3059
slr0093-5UTR (5'UTR) - TU3059
TU3059 (slr0093, slr0095) - Transcr Unit
TU3409 (sll0897, ssl1707, sll0898) - Transcr Unit
dnaK/sll1932 (mRNA / CDS) - TU588
dnaK/sll1932 (Protein) - TU588
TU588 (sll1932, sll1933) - Transcr Unit
dnaK1/sll0058 (mRNA / CDS) - TU2721
dnaK1/sll0058 (Protein) - TU2721
dnaK2/sll0170 (mRNA / CDS) - TU2403
dnaK2/sll0170 (Protein) - TU2403
TU2403 (sll0170) - Transcr Unit
sll0170-3UTR (3'UTR) - TU2403
sll0170-5UTR (5'UTR) - TU2403
dnaN/slr0965 (mRNA / CDS) - TU323
dnaN/slr0965 (Protein) - TU323
TU323 (slr0965) - Transcr Unit
dnaX/sll1360 (mRNA / CDS) - TU1095
dnaX/sll1360 (Protein) - TU1095
TU1095 (sll1360) - Transcr Unit
drgA/slr1719 (mRNA / CDS) - TU1506
drgA/slr1719 (Protein) - TU1506
dspA, dfr, hik33, nblS/sll0698 (mRNA / CDS) - TU112
dspA, dfr, hik33, nblS/sll0698 (Protein) - TU112
TU112 (sll0698, sll0699, sll0700) - Transcr Unit
dxr/sll0019 (mRNA / CDS) - TU2571
dxr/sll0019 (Protein) - TU2571
dxs/sll1945 (mRNA / CDS) - TU542
dxs/sll1945 (Protein) - TU542
sll1945-5UTR (5'UTR) - TU542
ecaB/slr0051 (mRNA / CDS) - TU2702
ecaB/slr0051 (Protein) - TU2702
TU2702 (slr0051) - Transcr Unit
efp/slr0434 (mRNA / CDS) - TU2674
efp/slr0434 (Protein) - TU2674
TU2674 (slr0434, slr0435, slr0436) - Transcr Unit
engA/slr1974 (mRNA / CDS) - TU1857
engA/slr1974 (Protein) - TU1857
eno/slr0752 (mRNA / CDS) - TU3597
eno/slr0752 (Protein) - TU3597
slr0752-3UTR (3'UTR) - TU3597
envM/slr1051 (mRNA / CDS) - TU490
envM/slr1051 (Protein) - TU490
epsB/sll0923 (mRNA / CDS) - TU344
epsB/sll0923 (Protein) - TU344
sll0923-3UTR (3'UTR) - TU344
sll0923-5UTR (5'UTR) - TU344
era/slr0321 (mRNA / CDS)
era/slr0321 (Protein)
erg6/slr0089 (mRNA / CDS) - TU3055
erg6/slr0089 (Protein) - TU3055
ETR1/slr1212 (mRNA / CDS)
ETR1/slr1212 (Protein)
exbB/slr0677 (mRNA / CDS) - TU393
exbB/slr0677 (Protein) - TU393
exoD/slr1875 (Protein) - TU1254
exsB/slr0527 (mRNA / CDS) - TU3346
exsB/slr0527 (Protein) - TU3346
fabD/slr2023 (mRNA / CDS) - TU769
fabD/slr2023 (Protein) - TU769
TU769 (slr2023) - Transcr Unit
fabF/sll1069 (mRNA / CDS) - TU823
fabF/sll1069 (Protein) - TU823
fabG/slr0886 (mRNA / CDS)
fabG/slr0886 (Protein)
fabH/slr1511 (Protein) - TU1638
TU1638 (slr1511) - Transcr Unit
fabZ/sll1605 (mRNA / CDS) - TU1503
fabZ/sll1605 (Protein) - TU1503
fbaA, fda/sll0018 (mRNA / CDS) - TU2572
fbaA, fda/sll0018 (Protein) - TU2572
sll0018-3UTR (3'UTR) - TU2572
sll0018-5UTR (5'UTR) - TU2572
TU2572 (sll0018) - Transcr Unit
fbp/sll1636 (mRNA / CDS) - TU527
fbp/sll1636 (Protein) - TU527
fbpI/slr2094 (mRNA / CDS) - TU1599
fbpI/slr2094 (Protein) - TU1599
TU1599 (slr2094) - Transcr Unit
slr2094-3UTR (3'UTR) - TU1599
fbpII/slr0952 (mRNA / CDS) - TU2095
fbpII/slr0952 (Protein) - TU2095
TU2095 (slr0952, slr0953, ncr0960) - Transcr Unit
fcl/sll1213 (mRNA / CDS) - TU9
fcl/sll1213 (Protein) - TU9
fda/slr0943 (mRNA / CDS) - TU2080
fda/slr0943 (Protein) - TU2080
TU2080 (slr0943) - Transcr Unit
fecB/slr1492 (Protein)
ffh/slr1531 (mRNA / CDS) - TU1970
ffh/slr1531 (Protein) - TU1970
TU1970 (slr1531) - Transcr Unit
slr1531-3UTR (3'UTR) - TU1970
ffs/ncr0810 (ncRNA) - TU1905
TU1905 (ffs, slr1474) - Transcr Unit
fmt/slr0070 (mRNA / CDS) - TU2737
fmt/slr0070 (Protein) - TU2737
fni/sll1556 (mRNA / CDS) - TU2050
fni/sll1556 (Protein) - TU2050
folC/sll1612 (mRNA / CDS) - TU1496
folC/sll1612 (Protein) - TU1496
folD/sll0753 (mRNA / CDS) - TU2514
folD/sll0753 (Protein) - TU2514
TU2514 (sll0753, ssl1417, sll0754, sll0755, sll0756) - Transcr Unit
folE/slr0426 (mRNA / CDS) - TU2873
folE/slr0426 (Protein) - TU2873
TU2873 (slr0426, ncr1395, slr0427) - Transcr Unit
TU181 (slr1093, ncr0090) - Transcr Unit
folP/slr2026 (mRNA / CDS) - TU775
folP/slr2026 (Protein) - TU775
TU775 (slr2026) - Transcr Unit
fpg/slr1689 (mRNA / CDS) - TU2056
fpg/slr1689 (Protein) - TU2056
fraH/slr0298 (mRNA / CDS) - TU3174
fraH/slr0298 (Protein) - TU3174
frmA/sll0990 (mRNA / CDS) - TU641
frmA/sll0990 (Protein) - TU641
frpC/sll1009 (mRNA / CDS) - TU491
frpC/sll1009 (Protein) - TU491
frr, rrf/sll0145 (mRNA / CDS) - TU2277
frr, rrf/sll0145 (Protein) - TU2277
ftrC/sll0554 (mRNA / CDS) - TU2948
ftrC/sll0554 (Protein) - TU2948
TU2948 (sll0554, sll0555, 6803t35, ncl1410, sll0556) - Transcr Unit
ftrV/ssr0330 (mRNA / CDS) - TU2907
ftrV/ssr0330 (Protein) - TU2907
TU2907 (ssr0330, slr0209) - Transcr Unit
ftsH/sll1463 (mRNA / CDS) - TU1008
ftsH/sll1463 (Protein) - TU1008
sll1463-5UTR (5'UTR) - TU1008
ftsH/slr0228 (mRNA / CDS) - TU2652
ftsH/slr0228 (Protein) - TU2652
TU2652 (slr0228) - Transcr Unit
slr0228-3UTR (3'UTR) - TU2652
ftsH/slr1390 (mRNA / CDS) - TU685
ftsH/slr1390 (Protein) - TU685
slr1390-5UTR (5'UTR) - TU685
ftsH/slr1604 (mRNA / CDS) - TU461
ftsH/slr1604 (Protein) - TU461
TU461 (slr1604) - Transcr Unit
slr1604-3UTR (3'UTR) - TU461
slr1604-5UTR (5'UTR) - TU461
TU1008 (sll1463, sll1464) - Transcr Unit
TU685 (slr1390, slr1391) - Transcr Unit
ftsY/slr2102 (mRNA / CDS) - TU1607
ftsY/slr2102 (Protein) - TU1607
slr2102-5UTR (5'UTR) - TU1607
TU1607 (slr2102, slr2103, ncr0610) - Transcr Unit
ftsZ/sll1633 (mRNA / CDS) - TU1031
ftsZ/sll1633 (Protein) - TU1031
TU1031 (sll1633) - Transcr Unit
sll1633-5UTR (5'UTR) - TU1031
fumC/slr0018 (mRNA / CDS)
fumC/slr0018 (Protein)
fur/sll0567 (mRNA / CDS) - TU3700
fur/sll0567 (Protein) - TU3700
TU3700 (sll0567) - Transcr Unit
fur/sll1937 (mRNA / CDS) - TU569
fur/sll1937 (Protein) - TU569
fus/sll0830 (mRNA / CDS) - TU2998
fus/sll0830 (Protein) - TU2998
TU2998 (sll0830) - Transcr Unit
fus/sll1098 (mRNA / CDS) - TU1986
fus/sll1098 (Protein) - TU1986
fus/slr1105 (mRNA / CDS) - TU195
fus/slr1105 (Protein) - TU195
slr1105-3UTR (3'UTR) - TU195
slr1105-5UTR (5'UTR) - TU195
fus/slr1463 (mRNA / CDS) - TU3492
fus/slr1463 (Protein) - TU3492
futA1/slr1295 (mRNA / CDS) - TU288
futA1/slr1295 (Protein) - TU288
TU288 (slr1295) - Transcr Unit
slr1295-3UTR (3'UTR) - TU288
futA2/slr0513 (mRNA / CDS) - TU3085
futA2/slr0513 (Protein) - TU3085
slr0513-3UTR (3'UTR) - TU3085
gabD/slr0370 (mRNA / CDS) - TU2456
gabD/slr0370 (Protein) - TU2456
slr0370-5UTR (5'UTR) - TU2456
gad/sll1641 (mRNA / CDS) - TU520
gad/sll1641 (Protein) - TU520
galE/sll0244 (mRNA / CDS) - TU1561
galE/sll0244 (Protein) - TU1561
galE/slr1067 (mRNA / CDS) - TU355
galE/slr1067 (Protein) - TU355
gap1/slr0884 (mRNA / CDS) - TU1187
gap1/slr0884 (Protein) - TU1187
TU1187 (slr0884, ncr0460) - Transcr Unit
gap2/sll1342 (mRNA / CDS) - TU3443
gap2/sll1342 (Protein) - TU3443
TU3443 (sll1342) - Transcr Unit
sll1342-3UTR (3'UTR) - TU3443
gatA/slr0877 (mRNA / CDS) - TU1171
gatA/slr0877 (Protein) - TU1171
gatB/sll1435 (mRNA / CDS) - TU1972
gatB/sll1435 (Protein) - TU1972
gatC/slr0033 (mRNA / CDS) - TU3318
gatC/slr0033 (Protein) - TU3318
slr0033-3UTR (3'UTR) - TU3318
gcvH/slr0879 (mRNA / CDS) - TU1174
gcvH/slr0879 (Protein) - TU1174
gcvP/slr0293 (mRNA / CDS) - TU2213
gcvP/slr0293 (Protein) - TU2213
gcvT/sll0171 (mRNA / CDS) - TU2402
gcvT/sll0171 (Protein) - TU2402
gdh/sll1709 (mRNA / CDS) - TU1310
gdh/sll1709 (Protein) - TU1310
gdhA/slr0710 (mRNA / CDS) - TU3283
gdhA/slr0710 (Protein) - TU3283
slr0710-5UTR (5'UTR) - TU3283
TU3283 (slr0710, slr0711, slr0712) - Transcr Unit
gdhB/slr1608 (mRNA / CDS) - TU473
gdhB/slr1608 (Protein) - TU473
slr1608-3UTR (3'UTR) - TU473
slr1608-as (asRNA)
slr1608-as2 (asRNA)
ggpS/sll1566 (mRNA / CDS) - TU2018
ggpS/sll1566 (Protein) - TU2018
TU2018 (sll1566) - Transcr Unit
sll1566-5UTR (5'UTR) - TU2018
ggt/slr1269 (mRNA / CDS) - TU1938
ggt/slr1269 (Protein) - TU1938
ggtA/slr0747 (mRNA / CDS) - TU3588
ggtA/slr0747 (Protein) - TU3588
TU3588 (slr0747) - Transcr Unit
ggtB/slr0529 (mRNA / CDS)
ggtB/slr0529 (Protein)
gidA/sll0202 (mRNA / CDS) - TU2643
gidA/sll0202 (Protein) - TU2643
gidA/sll0204 (mRNA / CDS) - TU2639
gidA/sll0204 (Protein) - TU2639
gidB/slr0072 (mRNA / CDS) - TU3025
gidB/slr0072 (Protein) - TU3025
TU3025 (slr0072) - Transcr Unit
TU627 (ssl1911) - Transcr Unit
gifB/sll1515 (mRNA / CDS) - TU441
gifB/sll1515 (Protein) - TU441
TU441 (sll1515) - Transcr Unit
sll1515-3UTR (3'UTR) - TU441
sll1515-5UTR (5'UTR) - TU441
glbN/slr2097 (mRNA / CDS) - TU1601
glbN/slr2097 (Protein) - TU1601
TU1601 (slr2097) - Transcr Unit
slr2097-3UTR (3'UTR) - TU1601
glcD/sll0404 (mRNA / CDS) - TU2681
glcD/sll0404 (Protein) - TU2681
TU2681 (sll0404) - Transcr Unit
sll0404-3UTR (3'UTR) - TU2681
glcE/sll1189 (mRNA / CDS) - TU305
glcE/sll1189 (Protein) - TU305
TU305 (sll1189) - Transcr Unit
glcF/sll1831 (mRNA / CDS) - TU618
glcF/sll1831 (Protein) - TU618
glcP, gtr/sll0771 (mRNA / CDS) - TU2495
glcP, gtr/sll0771 (Protein) - TU2495
sll0771-5UTR (5'UTR) - TU2495
TU2495 (sll0771) - Transcr Unit
glgA/sll0945 (mRNA / CDS) - TU2359
glgA/sll0945 (Protein) - TU2359
glgA/sll1393 (mRNA / CDS) - TU52
glgA/sll1393 (Protein) - TU52
TU52 (sll1393) - Transcr Unit
TU53 (sll1393) - Transcr Unit
sll1393-5UTR (5'UTR) - TU52
TU2359 (sll0945, 6803t27) - Transcr Unit
glgB/sll0158 (mRNA / CDS) - TU2422
glgB/sll0158 (Protein) - TU2422
TU2422 (sll0158, sll0160) - Transcr Unit
glgC/slr1176 (mRNA / CDS) - TU3679
glgC/slr1176 (Protein) - TU3679
glgP/sll1356 (mRNA / CDS) - TU1101
glgP/sll1356 (Protein) - TU1101
glgP/slr1367 (mRNA / CDS) - TU1150
glgP/slr1367 (Protein) - TU1150
slr1367-5UTR (5'UTR) - TU1150
glgX/slr0237 (mRNA / CDS) - TU122
glgX/slr0237 (Protein) - TU122
glgX/slr1857 (mRNA / CDS) - TU1227
glgX/slr1857 (Protein) - TU1227
glk/sll0593 (mRNA / CDS) - TU3108
glk/sll0593 (Protein) - TU3108
glmS/sll0220 (mRNA / CDS) - TU152
glmS/sll0220 (Protein) - TU152
sll0220-5UTR (5'UTR) - TU152
TU152 (sll0220, sll0221) - Transcr Unit
glmU/sll0899 (mRNA / CDS) - TU3405
glmU/sll0899 (Protein) - TU3405
glnA/slr1756 (mRNA / CDS) - TU510
glnA/slr1756 (Protein) - TU510
TU510 (slr1756) - Transcr Unit
slr1756-3UTR (3'UTR) - TU510
glnB/ssl0707 (mRNA / CDS) - TU2231
glnB/ssl0707 (Protein) - TU2231
TU2231 (ssl0707) - Transcr Unit
ssl0707-3UTR (3'UTR) - TU2231
ssl0707-5UTR (5'UTR) - TU2231
glnN/slr0288 (mRNA / CDS) - TU2207
glnN/slr0288 (Protein) - TU2207
TU2207 (slr0288, slr0291) - Transcr Unit
gloA/slr0381 (mRNA / CDS) - TU2472
gloA/slr0381 (Protein) - TU2472
slr0381-3UTR (3'UTR) - TU2472
glpD/sll1085 (mRNA / CDS) - TU2016
glpD/sll1085 (Protein) - TU2016
TU2016 (sll1085) - Transcr Unit
sll1085-3UTR (3'UTR) - TU2016
sll1085-5UTR (5'UTR) - TU2016
glpK/slr1672 (mRNA / CDS) - TU2022
glpK/slr1672 (Protein) - TU2022
TU2022 (slr1672, slr1673) - Transcr Unit
glsA/slr2079 (mRNA / CDS) - TU1570
glsA/slr2079 (Protein) - TU1570
glsF/sll1499 (mRNA / CDS) - TU3533
glsF/sll1499 (Protein) - TU3533
TU3533 (sll1499) - Transcr Unit
sll1499-3UTR (3'UTR) - TU3533
sll1499-5UTR (5'UTR) - TU3533
gltA/sll0401 (mRNA / CDS) - TU2692
gltA/sll0401 (Protein) - TU2692
gltB/sll1502 (mRNA / CDS) - TU467
gltB/sll1502 (Protein) - TU467
TU467 (sll1502) - Transcr Unit
gltD/sll1027 (mRNA / CDS) - TU209
gltD/sll1027 (Protein) - TU209
TU209 (sll1027) - Transcr Unit
gltX/sll0179 (mRNA / CDS) - TU2900
gltX/sll0179 (Protein) - TU2900
TU2900 (sll0179) - Transcr Unit
glyA/sll1931 (mRNA / CDS) - TU590
glyA/sll1931 (Protein) - TU590
TU590 (sll1931) - Transcr Unit
sll1931-3UTR (3'UTR) - TU590
glyQ/slr0638 (mRNA / CDS) - TU2811
glyQ/slr0638 (Protein) - TU2811
glyS/slr0220 (mRNA / CDS) - TU2636
glyS/slr0220 (Protein) - TU2636
TU2636 (slr0220) - Transcr Unit
gmhA/sll0083 (mRNA / CDS) - TU3036
gmhA/sll0083 (Protein) - TU3036
gmk/slr1123 (mRNA / CDS)
gmk/slr1123 (Protein)
gnd/sll0329 (mRNA / CDS) - TU2480
gnd/sll0329 (Protein) - TU2480
gpmB/slr1124 (mRNA / CDS) - TU80
gpmB/slr1124 (Protein) - TU80
gpsA/slr1755 (mRNA / CDS) - TU508
gpsA/slr1755 (Protein) - TU508
gpx1/slr1171 (mRNA / CDS) - TU2011
gpx1/slr1171 (Protein) - TU2011
TU2011 (slr1171, smr0014) - Transcr Unit
gpx2/slr1992 (mRNA / CDS) - TU1477
gpx2/slr1992 (Protein) - TU1477
TU1477 (slr1992) - Transcr Unit
slr1992-3UTR (3'UTR) - TU1477
groEL1/slr2076 (mRNA / CDS) - TU924
groEL1/slr2076 (Protein) - TU924
groEL-2/sll0416 (mRNA / CDS) - TU2663
groEL-2/sll0416 (Protein) - TU2663
sll0416-5UTR (5'UTR) - TU2663
TU2663 (sll0416, ncl1280) - Transcr Unit
groES/slr2075 (mRNA / CDS) - TU924
groES/slr2075 (Protein) - TU924
slr2075-5UTR (5'UTR) - TU924
TU924 (slr2075, slr2076) - Transcr Unit
grpE/sll0057 (mRNA / CDS) - TU2721
grpE/sll0057 (Protein) - TU2721
TU2721 (sll0057, sll0058, sll0060) - Transcr Unit
grxC/slr1562 (mRNA / CDS) - TU3549
grxC/slr1562 (Protein) - TU3549
grxC/ssr2061 (mRNA / CDS) - TU1083
grxC/ssr2061 (Protein) - TU1083
gshB/slr1238 (mRNA / CDS) - TU1083
gshB/slr1238 (Protein) - TU1083
gspD/slr1277 (mRNA / CDS) - TU1946
gspD/slr1277 (Protein) - TU1946
slr1277-3UTR (3'UTR) - TU1946
slr1277-5UTR (5'UTR) - TU1946
slr1277-as3 (asRNA)
gspE/slr0079 (mRNA / CDS) - TU3039
gspE/slr0079 (Protein) - TU3039
gst/sll0067 (mRNA / CDS) - TU2706
gst/sll0067 (Protein) - TU2706
gst1/sll1545 (mRNA / CDS) - TU3647
gst1/sll1545 (Protein) - TU3647
TU1985 (sll1102, sll1103, sll1104) - Transcr Unit
gtrB/sll1103 (mRNA / CDS) - TU1985
gtrB/sll1103 (Protein) - TU1985
gtrC/sll1104 (mRNA / CDS) - TU1985
gtrC/sll1104 (Protein) - TU1985
guaA/slr0213 (mRNA / CDS) - TU2615
guaA/slr0213 (Protein) - TU2615
slr0213-5UTR (5'UTR) - TU2615
slr0213-as (asRNA)
TU2615 (slr0213, slr0214) - Transcr Unit
guaB/slr0172 (mRNA / CDS) - TU2395
guaB/slr0172 (Protein) - TU2395
guaB/slr1722 (mRNA / CDS) - TU1508
guaB/slr1722 (Protein) - TU1508
slr1722-3UTR (3'UTR) - TU1508
gumB/sll1581 (mRNA / CDS) - TU707
gumB/sll1581 (Protein) - TU707
gyrA/slr0417 (mRNA / CDS) - TU2836
gyrA/slr0417 (Protein) - TU2836
slr0417-5UTR (5'UTR) - TU2836
TU2836 (slr0417, slr0418) - Transcr Unit
gyrB/sll2005 (mRNA / CDS) - TU1298
gyrB/sll2005 (Protein) - TU1298
HAGH1/sll1019 (mRNA / CDS)
HAGH1/sll1019 (Protein)
TU2259 (slr0143) - Transcr Unit
TU2260 (slr0143) - Transcr Unit
hdrB/slr0201 (mRNA / CDS) - TU2891
hdrB/slr0201 (Protein) - TU2891
hemA/slr1808 (mRNA / CDS) - TU1319
hemA/slr1808 (Protein) - TU1319
TU1319 (slr1808) - Transcr Unit
hemB/sll1994 (mRNA / CDS) - TU1681
hemB/sll1994 (Protein) - TU1681
hemC/slr1887 (mRNA / CDS) - TU1281
hemC/slr1887 (Protein) - TU1281
TU1281 (slr1887) - Transcr Unit
hemE/slr0536 (mRNA / CDS) - TU3363
hemE/slr0536 (Protein) - TU3363
slr0536-as2 (asRNA)
hemF/sll1185 (mRNA / CDS) - TU312
hemF/sll1185 (Protein) - TU312
hemH, scpA/slr0839 (mRNA / CDS) - TU3004
hemH, scpA/slr0839 (Protein) - TU3004
slr0839-3UTR (3'UTR) - TU3004
slr0839-5UTR (5'UTR) - TU3004
TU3004 (slr0839) - Transcr Unit
hemK/sll1237 (mRNA / CDS) - TU1689
hemK/sll1237 (Protein) - TU1689
TU1689 (sll1237) - Transcr Unit
sll1237-5UTR (5'UTR) - TU1689
hemL/sll0017 (mRNA / CDS) - TU2574
hemL/sll0017 (Protein) - TU2574
TU2574 (sll0017) - Transcr Unit
hflX/slr1521 (mRNA / CDS) - TU1654
hflX/slr1521 (Protein) - TU1654
hfq/ssr3341 (mRNA / CDS) - TU1845
hfq/ssr3341 (Protein) - TU1845
hglK/slr1519 (mRNA / CDS) - TU1653
hglK/slr1519 (Protein) - TU1653
hhoA/sll1679 (mRNA / CDS) - TU225
hhoA/sll1679 (Protein) - TU225
sll1679-5UTR (5'UTR) - TU225
TU225 (sll1679, sll1680, sll1681) - Transcr Unit
hhoB/sll1427 (mRNA / CDS) - TU1623
hhoB/sll1427 (Protein) - TU1623
sll1427-3UTR (3'UTR) - TU1623
sll1427-5UTR (5'UTR) - TU1623
hik1/slr1393 (mRNA / CDS) - TU691
hik1/slr1393 (Protein) - TU691
TU691 (slr1393) - Transcr Unit
TU692 (slr1393) - Transcr Unit
TU3355 (slr0533) - Transcr Unit
hik11/slr1414 (mRNA / CDS) - TU170
hik11/slr1414 (Protein) - TU170
slr1414-5UTR (5'UTR) - TU170
TU170 (slr1414, slr1415) - Transcr Unit
hik12/sll1672 (mRNA / CDS) - TU236
hik12/sll1672 (Protein) - TU236
TU236 (sll1672, sll1673) - Transcr Unit
hik13/sll1003 (mRNA / CDS) - TU501
hik13/sll1003 (Protein) - TU501
TU501 (sll1003) - Transcr Unit
sll1003-5UTR (5'UTR) - TU501
TU1116 (sll1353) - Transcr Unit
hik17/sll1687 (mRNA / CDS) - TU1341
hik17/sll1687 (Protein) - TU1341
hik19/sll1905 (mRNA / CDS) - TU1480
hik19/sll1905 (Protein) - TU1480
TU1480 (sll1905) - Transcr Unit
TU1525 (sll1590) - Transcr Unit
hik21/slr2098 (mRNA / CDS) - TU1602
hik21/slr2098 (Protein) - TU1602
TU1602 (slr2098) - Transcr Unit
TU1608 (slr2104) - Transcr Unit
hik23/slr1324 (mRNA / CDS)
hik23/slr1324 (Protein)
hik24/slr1969 (mRNA / CDS) - TU1840
hik24/slr1969 (Protein) - TU1840
TU1840 (slr1969) - Transcr Unit
TU1842 (slr1969) - Transcr Unit
slr1969-5UTR (5'UTR) - TU1840
slr1969-as (asRNA)
hik25/slr0222 (mRNA / CDS) - TU2641
hik25/slr0222 (Protein) - TU2641
TU2641 (slr0222) - Transcr Unit
hik26/slr0484 (mRNA / CDS) - TU2761
hik26/slr0484 (Protein) - TU2761
TU2761 (slr0484) - Transcr Unit
hik27/slr0640 (mRNA / CDS) - TU2811
hik27/slr0640 (Protein) - TU2811
TU3091 (sll0474) - Transcr Unit
sll1475 (mRNA / CDS) - TU3579
hik32/sll1475 (Protein) - TU3579
hik36/slr0073 (mRNA / CDS) - TU3026
hik36/slr0073 (Protein) - TU3026
TU3026 (slr0073) - Transcr Unit
slr0073-5UTR (5'UTR) - TU3026
hik37/sll0094 (mRNA / CDS) - TU3154
hik37/sll0094 (Protein) - TU3154
sll0094-5UTR (5'UTR) - TU3154
TU3154 (sll0094, sll0095) - Transcr Unit
hik38/slr1400 (mRNA / CDS) - TU386
hik38/slr1400 (Protein) - TU386
TU386 (slr1400, slr0670) - Transcr Unit
hik39/sll1296 (mRNA / CDS) - TU679
hik39/sll1296 (Protein) - TU679
hik4/sll1228 (mRNA / CDS) - TU1706
hik4/sll1228 (Protein) - TU1706
TU1706 (sll1228) - Transcr Unit
sll1228-5UTR (5'UTR) - TU1706
slr2099 (mRNA / CDS) - TU1604
hik40/slr2099 (Protein) - TU1604
hik41/sll1229 (mRNA / CDS) - TU1703
hik41/sll1229 (Protein) - TU1703
TU1703 (sll1229) - Transcr Unit
TU1704 (sll1229) - Transcr Unit
TU1705 (sll1229) - Transcr Unit
hik42/sll1555 (mRNA / CDS) - TU2052
hik42/sll1555 (Protein) - TU2052
TU2052 (sll1555) - Transcr Unit
hik43/slr0322 (mRNA / CDS) - TU2368
hik43/slr0322 (Protein) - TU2368
TU2368 (slr0322, slr0323, slr0324, slr0325) - Transcr Unit
hik5/sll1888 (mRNA / CDS) - TU1849
hik5/sll1888 (Protein) - TU1849
hik6/sll1871 (mRNA / CDS) - TU1874
hik6/sll1871 (Protein) - TU1874
TU1874 (sll1871) - Transcr Unit
hik8, sasA/sll0750 (mRNA / CDS) - TU2518
hik8, sasA/sll0750 (Protein) - TU2518
sll0750-as (asRNA)
slr0210 (mRNA / CDS) - TU2909
hik9/slr0210 (Protein) - TU2909
TU2909 (slr0210) - Transcr Unit
hisA/slr0652 (mRNA / CDS) - TU3499
hisA/slr0652 (Protein) - TU3499
hisB/sll0084 (Protein) - TU3035
hisB/slr0500 (mRNA / CDS) - TU3064
hisB/slr0500 (Protein) - TU3064
TU3064 (slr0500) - Transcr Unit
hisC/sll1713 (mRNA / CDS) - TU986
hisC/sll1713 (Protein) - TU986
hisC/sll1958 (mRNA / CDS) - TU1451
hisC/sll1958 (Protein) - TU1451
sll1958-5UTR (5'UTR) - TU1451
TU1451 (sll1958, sll1959) - Transcr Unit
hisD/slr0682 (mRNA / CDS) - TU401
hisD/slr0682 (Protein) - TU401
TU401 (slr0682) - Transcr Unit
slr0682-3UTR (3'UTR) - TU401
slr0682-5UTR (5'UTR) - TU401
hisD/slr1848 (mRNA / CDS) - TU534
hisD/slr1848 (Protein) - TU534
hisF/sll1893 (mRNA / CDS) - TU1832
hisF/sll1893 (Protein) - TU1832
hisG/sll0900 (mRNA / CDS) - TU3405
hisG/sll0900 (Protein) - TU3405
hisH/slr0084 (mRNA / CDS) - TU3049
hisH/slr0084 (Protein) - TU3049
TU3049 (slr0084) - Transcr Unit
hisIE/slr0608 (mRNA / CDS) - TU3716
hisIE/slr0608 (Protein) - TU3716
TU3716 (slr0608) - Transcr Unit
hisS/slr0357 (mRNA / CDS) - TU2558
hisS/slr0357 (Protein) - TU2558
TU2558 (slr0357) - Transcr Unit
hisS/slr1560 (mRNA / CDS) - TU3549
hisS/slr1560 (Protein) - TU3549
TU3549 (slr1560, slr1562, slr1563) - Transcr Unit
hitB/slr0327 (mRNA / CDS) - TU2370
hitB/slr0327 (Protein) - TU2370
TU690 (ssl2542, ncl0260) - Transcr Unit
TU1000 (ssr2595, slr1544, ncr0410) - Transcr Unit
hliC, scpB/ssl1633 (mRNA / CDS) - TU1176
hliC, scpB/ssl1633 (Protein) - TU1176
ssl1633-3UTR (3'UTR) - TU1176
ssl1633-5UTR (5'UTR) - TU1176
ssl1633-as (asRNA)
ssl1633-as2 (asRNA)
TU1176 (ssl1633) - Transcr Unit
hliD, scpE/ssr1789 (mRNA / CDS) - TU380
hliD, scpE/ssr1789 (Protein) - TU380
ssr1789-3UTR (3'UTR) - TU380
TU380 (ssr1789) - Transcr Unit
TU1647 (hliR1) - Transcr Unit
hlyB/sll1180 (mRNA / CDS) - TU1929
hlyB/sll1180 (Protein) - TU1929
sll1180-3UTR (3'UTR) - TU1929
sll1180-5UTR (5'UTR) - TU1929
hlyD/sll1181 (mRNA / CDS) - TU1927
hlyD/sll1181 (Protein) - TU1927
sll1181-3UTR (3'UTR) - TU1927
sll1181-5UTR (5'UTR) - TU1927
ho1/sll1184 (mRNA / CDS) - TU312
ho1/sll1184 (Protein) - TU312
sll1184-5UTR (5'UTR) - TU312
TU312 (sll1184, sll1185) - Transcr Unit
hoxE/sll1220 (mRNA / CDS) - TU1714
hoxE/sll1220 (Protein) - TU1714
hoxF/sll1221 (mRNA / CDS) - TU1714
hoxF/sll1221 (Protein) - TU1714
hoxH/sll1226 (mRNA / CDS) - TU1714
hoxH/sll1226 (Protein) - TU1714
hoxU/sll1223 (mRNA / CDS) - TU1714
hoxU/sll1223 (Protein) - TU1714
hoxY/sll1224 (mRNA / CDS) - TU1714
hoxY/sll1224 (Protein) - TU1714
hpcE/slr1877 (mRNA / CDS) - TU1260
hpcE/slr1877 (Protein) - TU1260
slr1877-5UTR (5'UTR) - TU1260
hsp33/sll1988 (mRNA / CDS) - TU1575
hsp33/sll1988 (Protein) - TU1575
TU1575 (sll1988) - Transcr Unit
sll1988-3UTR (3'UTR) - TU1575
sll1988-5UTR (5'UTR) - TU1575
hspA/sll1514 (mRNA / CDS) - TU445
hspA/sll1514 (Protein) - TU445
TU445 (sll1514) - Transcr Unit
sll1514-3UTR (3'UTR) - TU445
sll1514-5UTR (5'UTR) - TU445
htpG/sll0430 (mRNA / CDS)
htpG/sll0430 (Protein)
htrA/slr1204 (mRNA / CDS) - TU891
htrA/slr1204 (Protein) - TU891
slr1204-5UTR (5'UTR) - TU891
TU1296 (slr2135) - Transcr Unit
hypA1/slr1675 (mRNA / CDS) - TU2024
hypA1/slr1675 (Protein) - TU2024
hypB/sll1079 (mRNA / CDS) - TU795
hypB/sll1079 (Protein) - TU795
hypB/sll1432 (mRNA / CDS) - TU1978
hypB/sll1432 (Protein) - TU1978
TU1978 (sll1432, sll1433) - Transcr Unit
TU1817 (ssl3580, sll1900) - Transcr Unit
hypD/slr1498 (mRNA / CDS) - TU51
hypD/slr1498 (Protein) - TU51
TU51 (slr1498) - Transcr Unit
hypE/sll1462 (mRNA / CDS) - TU1010
hypE/sll1462 (Protein) - TU1010
IAP75/slr1227 (mRNA / CDS) - TU1055
IAP75/slr1227 (Protein) - TU1055
icd/slr1289 (mRNA / CDS) - TU275
icd/slr1289 (Protein) - TU275
TU275 (slr1289, slr1290) - Transcr Unit
icfA/slr1347 (mRNA / CDS) - TU1800
icfA/slr1347 (Protein) - TU1800
icfG/slr1860 (mRNA / CDS) - TU1227
icfG/slr1860 (Protein) - TU1227
ictB/slr1515 (mRNA / CDS) - TU1645
ictB/slr1515 (Protein) - TU1645
TU1645 (slr1515, ssr2551) - Transcr Unit
ileS/sll1362 (mRNA / CDS) - TU1092
ileS/sll1362 (Protein) - TU1092
TU1092 (sll1362) - Transcr Unit
sll1362-3UTR (3'UTR) - TU1092
ilvA/slr2072 (mRNA / CDS) - TU918
ilvA/slr2072 (Protein) - TU918
TU918 (slr2072) - Transcr Unit
ilvB/sll1981 (mRNA / CDS) - TU1592
ilvB/sll1981 (Protein) - TU1592
TU1592 (sll1981) - Transcr Unit
sll1981-3UTR (3'UTR) - TU1592
sll1981-5UTR (5'UTR) - TU1592
ilvC/sll1363 (mRNA / CDS) - TU3498
ilvC/sll1363 (Protein) - TU3498
TU3498 (sll1363) - Transcr Unit
sll1363-3UTR (3'UTR) - TU3498
ilvD/slr0452 (mRNA / CDS) - TU3672
ilvD/slr0452 (Protein) - TU3672
TU3672 (slr0452, slr0453) - Transcr Unit
ilvE/slr0032 (mRNA / CDS) - TU3317
ilvE/slr0032 (Protein) - TU3317
slr0032-5UTR (5'UTR) - TU3317
ilvG/slr2088 (mRNA / CDS) - TU1585
ilvG/slr2088 (Protein) - TU1585
slr2088-5UTR (5'UTR) - TU1585
TU1585 (slr2088, slr2089) - Transcr Unit
ilvN/sll0065 (mRNA / CDS) - TU2706
ilvN/sll0065 (Protein) - TU2706
TU2706 (sll0065, sll0066, sll0067) - Transcr Unit
im30/slr1188 (mRNA / CDS) - TU3686
im30/slr1188 (Protein) - TU3686
infA/ssl3441 (mRNA / CDS) - TU835
infA/ssl3441 (Protein) - TU835
TU835 (ssl3441) - Transcr Unit
ssl3441-3UTR (3'UTR) - TU835
ssl3441-5UTR (5'UTR) - TU835
infB/slr0744 (mRNA / CDS) - TU119
infB/slr0744 (Protein) - TU119
slr0744-as2 (asRNA)
infC/slr0974 (mRNA / CDS) - TU341
infC/slr0974 (Protein) - TU341
int/slr0819 (mRNA / CDS) - TU1756
int/slr0819 (Protein) - TU1754
TU3327 (isaR1) - Transcr Unit
isaR1/ncl1600 (ncRNA) - TU3327
TU1556 (sll0247) - Transcr Unit
isiB/sll0248 (Protein) - TU1555
TU1555 (sll0248, sll0249) - Transcr Unit
ispD/slr0951 (mRNA / CDS) - TU2094
ispD/slr0951 (Protein) - TU2094
ispE/sll0711 (mRNA / CDS) - TU97
ispE/sll0711 (Protein) - TU97
ispF/slr1542 (mRNA / CDS) - TU995
ispF/slr1542 (Protein) - TU995
ispG/slr2136 (mRNA / CDS) - TU1297
ispG/slr2136 (Protein) - TU1297
ispH/slr0348 (mRNA / CDS) - TU2538
ispH/slr0348 (Protein) - TU2538
kaiA/slr0756 (mRNA / CDS) - TU3611
kaiA/slr0756 (Protein) - TU3611
TU3611 (slr0756, slr0757, slr0758) - Transcr Unit
kaiB1/slr0757 (Protein) - TU3611
kaiB2/sll1596 (Protein)
kaiB3/sll0486 (Protein) - TU3068
kaiC1/slr0758 (mRNA / CDS) - TU3611
kaiC1/slr0758 (Protein) - TU3611
kaiC2/sll1595 (mRNA / CDS)
kaiC2/sll1595 (Protein)
kaiC3/slr1942 (mRNA / CDS) - TU2333
kaiC3/slr1942 (Protein) - TU2333
TU2333 (slr1942) - Transcr Unit
kdpA/slr1728 (mRNA / CDS) - TU1526
kdpA/slr1728 (Protein) - TU1526
TU1526 (slr1728, slr1729, ssr2912, slr1730) - Transcr Unit
kdpB/slr1729 (mRNA / CDS) - TU1526
kdpB/slr1729 (Protein) - TU1526
kdpC/slr1730 (mRNA / CDS) - TU1526
kdpC/slr1730 (Protein) - TU1526
kdpD/slr1731 (mRNA / CDS)
kdpD/slr1731 (Protein)
slr1731-as (asRNA)
kdtB/slr0847 (Protein) - TU3023
kpsT/slr2108 (mRNA / CDS) - TU1668
kpsT/slr2108 (Protein) - TU1668
slr2108-3UTR (3'UTR) - TU1668
ksgA/sll0708 (Protein) - TU100
lacF/slr1202 (mRNA / CDS) - TU889
lacF/slr1202 (Protein) - TU889
lacG/slr1723 (Protein) - TU1509
slr1723-5UTR (5'UTR) - TU1509
lap/sll2001 (mRNA / CDS) - TU1660
lap/sll2001 (Protein) - TU1660
lepA/slr0604 (mRNA / CDS) - TU3711
lepA/slr0604 (Protein) - TU3711
slr0604-5UTR (5'UTR) - TU3711
slr0604-as2 (asRNA)
lepB/sll0716 (mRNA / CDS) - TU93
lepB/sll0716 (Protein) - TU93
sll0716-5UTR (5'UTR) - TU93
lepB/slr1377 (mRNA / CDS) - TU671
lepB/slr1377 (Protein) - TU671
leuA/sll1564 (mRNA / CDS) - TU2029
leuA/sll1564 (Protein) - TU2029
sll1564-5UTR (5'UTR) - TU2029
leuA/slr0186 (mRNA / CDS) - TU2432
leuA/slr0186 (Protein) - TU2432
slr0186-5UTR (5'UTR) - TU2432
TU2432 (slr0186, slr0359) - Transcr Unit
leuB/slr1517 (mRNA / CDS) - TU1650
leuB/slr1517 (Protein) - TU1650
TU1650 (slr1517) - Transcr Unit
leuC/sll1470 (mRNA / CDS) - TU3584
leuC/sll1470 (Protein) - TU3584
TU3584 (sll1470) - Transcr Unit
sll1470-3UTR (3'UTR) - TU3584
leuD/sll1444 (mRNA / CDS) - TU1962
leuD/sll1444 (Protein) - TU1962
leuS/sll1074 (mRNA / CDS) - TU805
leuS/sll1074 (Protein) - TU805
TU805 (sll1074, ssl2100) - Transcr Unit
lexA/sll1626 (mRNA / CDS) - TU1352
lexA/sll1626 (Protein) - TU1352
sll1626-3UTR (3'UTR) - TU1352
sll1626-5UTR (5'UTR) - TU1352
lig/sll1209 (mRNA / CDS) - TU15
lig/sll1209 (Protein) - TU15
lig/sll1583 (mRNA / CDS) - TU706
lig/sll1583 (Protein) - TU706
sll1583-5UTR (5'UTR) - TU706
lim17/sll0626 (mRNA / CDS) - TU3523
lim17/sll0626 (Protein) - TU3523
lipA/slr1598 (mRNA / CDS) - TU448
lipA/slr1598 (Protein) - TU448
lipB/slr0994 (mRNA / CDS)
lipB/slr0994 (Protein)
llaI.2/sll0709 (mRNA / CDS) - TU97
llaI.2/sll0709 (Protein) - TU97
lmbP/slr0862 (mRNA / CDS) - TU1395
lmbP/slr0862 (Protein) - TU1395
lpxA/sll0379 (Protein) - TU2847
lpxB/slr0015 (mRNA / CDS) - TU2592
lpxB/slr0015 (Protein) - TU2592
lpxC/sll1508 (mRNA / CDS)
lpxC/sll1508 (Protein)
lpxD/slr0776 (mRNA / CDS) - TU2497
lpxD/slr0776 (Protein) - TU2497
lrtA/sll0947 (mRNA / CDS) - TU2356
lrtA/sll0947 (Protein) - TU2356
lysA/sll0504 (mRNA / CDS) - TU3378
lysA/sll0504 (Protein) - TU3378
lysC/slr0657 (mRNA / CDS) - TU3516
lysC/slr0657 (Protein) - TU3516
lysS/slr1550 (mRNA / CDS) - TU1015
lysS/slr1550 (Protein) - TU1015
lytB/slr0191 (mRNA / CDS) - TU2876
lytB/slr0191 (Protein) - TU2876
maf/sll0905 (mRNA / CDS) - TU2103
maf/sll0905 (Protein) - TU2103
sll0905-as (asRNA)
malK/slr1224 (mRNA / CDS) - TU1049
malK/slr1224 (Protein) - TU1049
slr1224-as2 (asRNA)
malQ/sll1676 (mRNA / CDS) - TU231
malQ/sll1676 (Protein) - TU231
manA/slr2074 (mRNA / CDS) - TU920
manA/slr2074 (Protein) - TU920
map/slr0918 (mRNA / CDS) - TU2937
map/slr0918 (Protein) - TU2937
sll1005 (mRNA / CDS)
mazG/sll1005 (Protein)
me/slr0721 (mRNA / CDS) - TU95
me/slr0721 (Protein) - TU95
MelB/sll1374 (mRNA / CDS) - TU1918
MelB/sll1374 (Protein) - TU1918
menB/sll1127 (mRNA / CDS)
menB/sll1127 (Protein)
sll1127-as (asRNA)
sll1127-as2 (asRNA)
menD/sll0603 (mRNA / CDS) - TU2819
menD/sll0603 (Protein) - TU2819
TU1752 (slr0817) - Transcr Unit
menG/sll1653 (mRNA / CDS) - TU272
menG/sll1653 (Protein) - TU272
TU272 (sll1653) - Transcr Unit
metH/slr0212 (mRNA / CDS) - TU2913
metH/slr0212 (Protein) - TU2913
TU2913 (slr0212, ssr0335, ssr0336) - Transcr Unit
metS/slr0649 (mRNA / CDS) - TU2828
metS/slr0649 (Protein) - TU2828
TU2828 (slr0649, slr0650) - Transcr Unit
metX/sll0927 (mRNA / CDS) - TU335
metX/sll0927 (Protein) - TU335
mgtE/slr1216 (mRNA / CDS)
mgtE/slr1216 (Protein)
miaA/sll0817 (Protein) - TU1737
miaB/sll0996 (mRNA / CDS) - TU631
miaB/sll0996 (Protein) - TU631
minC/sll0288 (mRNA / CDS) - TU3173
minC/sll0288 (Protein) - TU3173
minD/sll0289 (mRNA / CDS) - TU3172
minD/sll0289 (Protein) - TU3172
TU3172 (sll0289, ssl0546) - Transcr Unit
minE/ssl0546 (mRNA / CDS) - TU3172
minE/ssl0546 (Protein) - TU3172
mltA/sll0016 (mRNA / CDS) - TU2576
mltA/sll0016 (Protein) - TU2576
sll0016-3UTR (3'UTR) - TU2576
sll0016-5UTR (5'UTR) - TU2576
mmsB/slr0229 (mRNA / CDS) - TU2657
mmsB/slr0229 (Protein) - TU2657
mnmA/sll0844 (mRNA / CDS) - TU1397
mnmA/sll0844 (Protein) - TU1397
sll0844-3UTR (3'UTR) - TU1397
sll0844-5UTR (5'UTR) - TU1397
moaD/ssr1527 (Protein) - TU2925
moaE/slr0903 (Protein) - TU2925
moeA/slr0900 (mRNA / CDS) - TU2925
moeA/slr0900 (Protein) - TU2925
moeB/sll1536 (mRNA / CDS) - TU2120
moeB/sll1536 (Protein) - TU2120
TU2120 (sll1536) - Transcr Unit
sll1536-3UTR (3'UTR) - TU2120
mom72/sll1667 (mRNA / CDS) - TU247
mom72/sll1667 (Protein) - TU247
morR/slr1416 (mRNA / CDS) - TU172
morR/slr1416 (Protein) - TU172
moxR/slr0835 (mRNA / CDS) - TU2994
moxR/slr0835 (Protein) - TU2994
mraY/sll0657 (mRNA / CDS) - TU3292
mraY/sll0657 (Protein) - TU3292
mrcA/sll1434 (mRNA / CDS) - TU1973
mrcA/sll1434 (Protein) - TU1973
sll1434-5UTR (5'UTR) - TU1973
mrcB/slr1710 (mRNA / CDS) - TU748
mrcB/slr1710 (Protein) - TU748
slr1710-5UTR (5'UTR) - TU748
mrp/slr0067 (mRNA / CDS) - TU2734
mrp/slr0067 (Protein) - TU2734
slr0067-5UTR (5'UTR) - TU2734
mscL/slr0875 (mRNA / CDS) - TU1163
mscL/slr0875 (Protein) - TU1163
slr0875-3UTR (3'UTR) - TU1163
msrA/sll1394 (mRNA / CDS) - TU50
msrA/sll1394 (Protein) - TU50
msrA/slr1795 (mRNA / CDS) - TU246
msrA/slr1795 (Protein) - TU246
mtfB/sll1231 (Protein)
mtnA/slr1938 (mRNA / CDS) - TU2324
mtnA/slr1938 (Protein) - TU2324
mtnP/sll0135 (mRNA / CDS) - TU2288
mtnP/sll0135 (Protein) - TU2288
murA/slr0017 (mRNA / CDS) - TU2598
murA/slr0017 (Protein) - TU2598
murB/slr1424 (mRNA / CDS) - TU3448
murB/slr1424 (Protein) - TU3448
murC/slr1423 (mRNA / CDS) - TU3448
murC/slr1423 (Protein) - TU3448
slr1423-5UTR (5'UTR) - TU3448
TU3446 (slr1423, slr1424, slr1425) - Transcr Unit
TU3447 (slr1423, slr1424, slr1425) - Transcr Unit
TU3448 (slr1423, slr1424, slr1425) - Transcr Unit
murD/sll2010 (mRNA / CDS) - TU1291
murD/sll2010 (Protein) - TU1291
TU1291 (sll2010, sll2011) - Transcr Unit
murE/slr0528 (mRNA / CDS) - TU3348
murE/slr0528 (Protein) - TU3348
TU3348 (slr0528) - Transcr Unit
slr0528-as (asRNA)
murF/slr1351 (mRNA / CDS) - TU1805
murF/slr1351 (Protein) - TU1805
TU1805 (slr1351) - Transcr Unit
slr1351-5UTR (5'UTR) - TU1805
murG/slr1656 (mRNA / CDS) - TU3634
murG/slr1656 (Protein) - TU3634
TU3634 (slr1656) - Transcr Unit
TU3635 (slr1656) - Transcr Unit
TU1372 (slr1746, ncr0520) - Transcr Unit
murQ/sll0861 (mRNA / CDS) - TU1200
murQ/sll0861 (Protein) - TU1200
mutS/sll1165 (mRNA / CDS) - TU1412
mutS/sll1165 (Protein) - TU1412
mutT/sll1045 (mRNA / CDS)
mutT/sll1045 (Protein)
6803t01 (tRNA)
6803t02 (tRNA) - TU241
6803t03 (tRNA) - TU256
6803t04 (tRNA) - TU317
6803t05 (tRNA) - TU342
6803t06 (tRNA) - TU430
6803t07 (tRNA) - TU629
6803t08 (tRNA) - TU641
6803t09 (tRNA) - TU934
6803t10 (tRNA) - TU1004
6803t11 (tRNA) - TU1111
6803t12 (tRNA) - TU1125
6803t13 (tRNA) - TU1364
6803t14 (tRNA) - TU1442
6803t15 (tRNA) - TU1587
6803t16 (tRNA) - TU1663
6803t17 (tRNA)
6803t18 (tRNA) - TU1780
6803t19 (tRNA) - TU1890
6803t20 (tRNA) - TU1904
6803t21 (tRNA)
6803t22 (tRNA) - TU2132
6803t23 (tRNA) - TU2168
6803t24 (tRNA)
6803t25 (tRNA)
6803t26 (tRNA)
6803t27 (tRNA) - TU2359
6803t28 (tRNA)
6803t29 (tRNA)
6803t30 (tRNA)
6803t31 (tRNA) - TU2613
6803t32 (tRNA) - TU2719
6803t33 (tRNA) - TU2792
6803t34 (tRNA) - TU2935
6803t35 (tRNA) - TU2948
6803t36 (tRNA)
6803t37 (tRNA) - TU3394
6803t38 (tRNA) - TU3394
6803t39 (tRNA) - TU3465
6803t40 (tRNA)
6803t41 (tRNA) - TU3514
6803t42 (tRNA)
ncl0060 (ncRNA) - TU201
ncl0070 (ncRNA) - TU205
ncl0100 (ncRNA) - TU331
ncl0110 (ncRNA) - TU332
ncl0150 (ncRNA) - TU463
ncl0160 (ncRNA) - TU475
ncl0170 (ncRNA) - TU491
ncl0200 (ncRNA) - TU530
ncl0215 (ncRNA) - TU581
ncl0260 (ncRNA) - TU690
ncl0270 (ncRNA) - TU715
ncl0310 (ncRNA) - TU766
ncl0320 (ncRNA) - TU784
ncl0350 (ncRNA) - TU795
ncl0370 (ncRNA) - TU830
ncl0380 (ncRNA) - TU837
ncl0400 (ncRNA) - TU908
ncl0430 (ncRNA)
ncl0490 (ncRNA) - TU1144
ncl0500 (ncRNA) - TU1186
ncl0530 (ncRNA) - TU1288
ncl0560 (ncRNA) - TU1338
ncl0570 (ncRNA) - TU1341
ncl0590 (ncRNA) - TU1388
ncl0620 (ncRNA) - TU1440
ncl0710 (ncRNA) - TU1593
ncl0800 (ncRNA) - TU1714
ncl0820 (ncRNA) - TU1737
ncl0845 (ncRNA) - TU1804
ncl0860 (ncRNA) - TU1829
ncl0940 (ncRNA)
ncl0980 (ncRNA) - TU2054
ncl1010 (ncRNA) - TU2125
ncl1020 (ncRNA) - TU2142
ncl1040 (ncRNA) - TU2166
ncl1045 (ncRNA) - TU2191
ncl1110 (ncRNA) - TU2356
ncl1120 (ncRNA) - TU2356
ncl1130 (ncRNA) - TU2366
ncl1150 (ncRNA) - TU2406
ncl1180 (ncRNA) - TU2474
ncl1190 (ncRNA) - TU2480
ncl1255 (ncRNA) - TU2629
ncl1260 (ncRNA) - TU2646
ncl1280 (ncRNA) - TU2663
ncl1310 (ncRNA) - TU2754
ncl1330 (ncRNA) - TU2824
ncl1350 (ncRNA) - TU2857
ncl1390 (ncRNA) - TU2884
ncl1410 (ncRNA) - TU2948
ncl1450 (ncRNA) - TU3007
ncl1460 (ncRNA) - TU3022
ncl1470 (ncRNA) - TU3100
ncl1490 (ncRNA) - TU3138
ncl1500 (ncRNA) - TU3138
ncl1570 (ncRNA) - TU3300
ncl1580 (ncRNA) - TU3301
ncl1590 (ncRNA) - TU3314
ncl1650 (ncRNA) - TU3414
ncl1690 (ncRNA) - TU3503
ncl1700 (ncRNA)
ncl1780 (ncRNA) - TU3698
ncl1790 (ncRNA) - TU3718
ncr0020 (ncRNA) - TU87
ncr0070 (ncRNA) - TU127
ncr0080 (ncRNA) - TU161
ncr0200 (ncRNA) - TU420
ncr0210 (ncRNA) - TU433
ncr0240 (ncRNA) - TU528
ncr0270 (ncRNA) - TU611
ncr0280 (ncRNA) - TU643
ncr0310 (ncRNA) - TU731
ncr0320 (ncRNA) - TU741
ncr0340 (ncRNA)
ncr0380 (ncRNA) - TU905
ncr0410 (ncRNA) - TU1000
ncr0420 (ncRNA) - TU1041
ncr0440 (ncRNA) - TU1103
ncr0450 (ncRNA) - TU1129
ncr0510 (ncRNA) - TU1360
ncr0520 (ncRNA) - TU1373
ncr0530 (ncRNA) - TU1454
ncr0560 (ncRNA) - TU1460
ncr0570 (ncRNA) - TU1471
ncr0575 (ncRNA) - TU1472
ncr0630 (ncRNA) - TU1628
ncr0650 (ncRNA) - TU1663
ncr0670 (ncRNA) - TU1695
ncr0710 (ncRNA) - TU1715
ncr0720 (ncRNA) - TU1748
ncr0730 (ncRNA) - TU1761
ncr0760 (ncRNA) - TU1847
ncr0800 (ncRNA) - TU1891
ncr0830 (ncRNA) - TU1945
ncr0840 (ncRNA) - TU1953
ncr0860 (ncRNA) - TU1957
ncr0890 (ncRNA) - TU2009
ncr0910 (ncRNA)
ncr0920 (ncRNA) - TU2027
ncr0960 (ncRNA) - TU2098
ncr0970 (ncRNA) - TU2144
ncr1010 (ncRNA) - TU2183
ncr1020 (ncRNA) - TU2205
ncr1071 (ncRNA) - TU2296
ncr1200 (ncRNA) - TU2463
ncr1220 (ncRNA) - TU2529
ncr1240 (ncRNA) - TU2555
ncr1280 (ncRNA) - TU2598
ncr1310 (ncRNA)
ncr1320 (ncRNA) - TU2657
ncr1350 (ncRNA) - TU2746
ncr1420 (ncRNA) - TU2899
ncr1450 (ncRNA) - TU2969
ncr1460 (ncRNA) - TU2986
ncr1500 (ncRNA) - TU3043
ncr1570 (ncRNA) - TU3330
ncr1580 (ncRNA) - TU3402
ncr1590 (ncRNA) - TU3417
ncr1600 (ncRNA) - TU3419
ncr1640 (ncRNA) - TU3492
ncr1650 (ncRNA) - TU3504
ncr1660 (ncRNA) - TU3510
ncr1680 (ncRNA) - TU3599
sll0005 (mRNA / CDS) - TU2606
sll0005 (Protein) - TU2606
sll0007 (mRNA / CDS) - TU2603
sll0007 (Protein) - TU2603
sll0008 (Protein) - TU2603
sll0012 (transp mRNA / CDS)
sll0021 (mRNA / CDS) - TU2569
sll0021 (Protein) - TU2569
sll0021-5UTR (5'UTR) - TU2569
sll0022 (mRNA / CDS) - TU2567
sll0022 (Protein) - TU2567
sll0023 (mRNA / CDS) - TU2567
sll0023 (Protein) - TU2567
sll0034 (mRNA / CDS) - TU3324
sll0034 (Protein) - TU3324
sll0036 (mRNA / CDS) - TU3321
sll0036 (Protein) - TU3321
sll0044 (mRNA / CDS) - TU3311
sll0044 (Protein) - TU3311
sll0048 (mRNA / CDS) - TU3301
sll0048 (Protein) - TU3301
sll0048-5UTR (5'UTR) - TU3301
sll0051 (mRNA / CDS) - TU2736
sll0051 (Protein) - TU2736
sll0051-3UTR (3'UTR) - TU2736
sll0051-5UTR (5'UTR) - TU2736
sll0055 (mRNA / CDS) - TU2733
sll0055 (Protein) - TU2733
sll0063 (mRNA / CDS) - TU2716
sll0063 (Protein) - TU2716
sll0064 (mRNA / CDS) - TU2714
sll0064 (Protein) - TU2714
sll0066 (mRNA / CDS) - TU2706
sll0066 (Protein) - TU2706
sll0068 (mRNA / CDS) - TU2704
sll0068 (Protein) - TU2704
sll0068-5UTR (5'UTR) - TU2704
sll0069 (mRNA / CDS) - TU2703
sll0069 (Protein) - TU2703
sll0069-5UTR (5'UTR) - TU2703
sll0072 (mRNA / CDS)
sll0072 (Protein)
sll0085 (mRNA / CDS) - TU3034
sll0085 (Protein) - TU3033
sll0086 (mRNA / CDS) - TU3031
sll0086 (Protein) - TU3031
sll0086-3UTR (3'UTR) - TU3031
sll0088 (mRNA / CDS) - TU3027
sll0088 (Protein) - TU3027
sll0088-5UTR (5'UTR) - TU3027
sll0092 (transp mRNA / CDS) - TU3163
sll0095 (mRNA / CDS) - TU3154
sll0095 (Protein) - TU3154
sll0096 (mRNA / CDS) - TU3153
sll0096 (Protein) - TU3153
sll0102 (mRNA / CDS) - TU3148
sll0102 (Protein) - TU3148
sll0103 (mRNA / CDS) - TU3147
sll0103 (Protein) - TU3147
sll0141 (mRNA / CDS) - TU2282
sll0141 (Protein) - TU2282
sll0142 (mRNA / CDS) - TU2281
sll0142 (Protein) - TU2281
sll0147 (mRNA / CDS) - TU2274
sll0147 (Protein) - TU2274
sll0148 (mRNA / CDS) - TU2272
sll0148 (Protein) - TU2272
sll0149 (mRNA / CDS) - TU2271
sll0149 (Protein) - TU2271
sll0154 (mRNA / CDS) - TU2431
sll0154 (Protein) - TU2431
sll0154-5UTR (5'UTR) - TU2431
sll0157 (mRNA / CDS) - TU2425
sll0157 (Protein) - TU2425
sll0160 (mRNA / CDS) - TU2422
sll0160 (Protein) - TU2422
sll0162 (mRNA / CDS)
sll0162 (Protein)
sll0163 (mRNA / CDS) - TU2411
sll0163 (Protein) - TU2411
sll0163-as (asRNA)
sll0167 (mRNA / CDS) - TU2408
sll0167 (Protein) - TU2408
sll0167-5UTR (5'UTR) - TU2408
sll0169 (mRNA / CDS) - TU2406
sll0169 (Protein) - TU2406
sll0172 (mRNA / CDS) - TU2398
sll0172 (Protein) - TU2398
sll0172-5UTR (5'UTR) - TU2398
sll0174 (mRNA / CDS) - TU2398
sll0174 (Protein) - TU2398
sll0175 (mRNA / CDS) - TU2398
sll0175 (Protein) - TU2398
sll0178 (mRNA / CDS) - TU2914
sll0178 (Protein) - TU2914
sll0180 (mRNA / CDS) - TU2896
sll0180 (Protein) - TU2896
sll0180-5UTR (5'UTR) - TU2896
sll0182 (mRNA / CDS) - TU2896
sll0182 (Protein) - TU2896
sll0183 (mRNA / CDS) - TU2893
sll0183 (Protein) - TU2893
sll0183-5UTR (5'UTR) - TU2893
sll0185 (mRNA / CDS) - TU2884
sll0185 (Protein) - TU2884
sll0188 (mRNA / CDS) - TU2874
sll0188 (Protein) - TU2874
sll0188-5UTR (5'UTR) - TU2874
sll0192 (mRNA / CDS) - TU2658
sll0192 (Protein) - TU2658
sll0198 (Protein) - TU2650
sll0200 (transp mRNA / CDS) - TU2644
sll0205 (mRNA / CDS) - TU2637
sll0205 (Protein) - TU2637
sll0208 (mRNA / CDS) - TU2628
sll0208 (Protein) - TU2628
sll0209 (mRNA / CDS) - TU2623
sll0209 (Protein) - TU2623
sll0216 (Protein) - TU164
sll0218 (mRNA / CDS) - TU156
sll0218 (Protein) - TU156
sll0224 (mRNA / CDS) - TU137
sll0224 (Protein) - TU137
sll0230 (mRNA / CDS) - TU128
sll0230 (Protein) - TU128
sll0236 (mRNA / CDS) - TU1569
sll0236 (Protein) - TU1569
sll0237 (mRNA / CDS) - TU1569
sll0237 (Protein) - TU1569
sll0242 (mRNA / CDS) - TU1567
sll0242 (Protein) - TU1567
sll0242-5UTR (5'UTR) - TU1567
sll0243 (mRNA / CDS) - TU1567
sll0243 (Protein) - TU1567
sll0245 (mRNA / CDS) - TU1559
sll0245 (Protein) - TU1559
sll0245-5UTR (5'UTR) - TU1559
sll0253 (mRNA / CDS) - TU1540
sll0253 (Protein) - TU1540
sll0253-3UTR (3'UTR) - TU1540
sll0254 (mRNA / CDS) - TU1538
sll0254 (Protein) - TU1538
sll0254-5UTR (5'UTR) - TU1538
sll0260 (mRNA / CDS) - TU2204
sll0260 (Protein) - TU2204
sll0263 (mRNA / CDS) - TU2195
sll0263 (Protein) - TU2195
sll0267 (mRNA / CDS) - TU2191
sll0267 (Protein) - TU2191
sll0268 (mRNA / CDS) - TU2191
sll0268 (Protein) - TU2191
sll0272 (mRNA / CDS) - TU2187
sll0272 (Protein) - TU2187
sll0274 (mRNA / CDS) - TU2186
sll0274 (Protein) - TU2186
sll0274-3UTR (3'UTR) - TU2186
sll0280 (mRNA / CDS)
sll0280 (Protein)
sll0281 (mRNA / CDS)
sll0281 (Protein)
sll0283 (mRNA / CDS) - TU3195
sll0283 (Protein) - TU3195
sll0284 (mRNA / CDS) - TU3192
sll0284 (Protein) - TU3192
sll0293 (mRNA / CDS) - TU2388
sll0293 (Protein) - TU2388
sll0295 (Protein) - TU2382
sll0296 (Protein) - TU2373
sll0301 (mRNA / CDS) - TU2369
sll0301 (Protein) - TU2369
sll0309 (Protein) - TU2565
sll0310 (mRNA / CDS)
sll0310 (Protein)
sll0312 (mRNA / CDS) - TU2557
sll0312 (Protein) - TU2557
sll0314 (mRNA / CDS) - TU2548
sll0314 (Protein) - TU2548
sll0314-5UTR (5'UTR) - TU2548
sll0315 (transp mRNA / CDS) - TU2544
sll0318 (mRNA / CDS) - TU2539
sll0318 (Protein) - TU2539
sll0319 (mRNA / CDS) - TU2535
sll0319 (Protein) - TU2535
sll0321 (mRNA / CDS) - TU2532
sll0321 (Protein) - TU2532
sll0325 (mRNA / CDS) - TU2524
sll0325 (Protein) - TU2524
sll0335 (mRNA / CDS)
sll0335 (Protein)
sll0354 (mRNA / CDS) - TU2238
sll0354 (Protein) - TU2238
sll0359 (mRNA / CDS) - TU2225
sll0359 (Protein) - TU2225
sll0359-3UTR (3'UTR) - TU2225
sll0361 (mRNA / CDS) - TU2221
sll0361 (Protein) - TU2221
sll0364 (Protein)
sll0376 (mRNA / CDS) - TU2855
sll0376 (Protein) - TU2855
sll0376-3UTR (3'UTR) - TU2855
sll0376-5UTR (5'UTR) - TU2855
sll0381 (mRNA / CDS) - TU2838
sll0381 (Protein) - TU2838
sll0394 (mRNA / CDS) - TU2832
sll0394 (Protein) - TU2832
sll0394-5UTR (5'UTR) - TU2832
sll0395 (mRNA / CDS) - TU2832
sll0395 (Protein) - TU2832
sll0396 (mRNA / CDS) - TU2831
sll0396 (Protein) - TU2831
sll0400 (mRNA / CDS) - TU2692
sll0400 (Protein) - TU2692
sll0400-5UTR (5'UTR) - TU2692
sll0405 (mRNA / CDS) - TU2676
sll0405 (Protein) - TU2676
sll0406 (mRNA / CDS) - TU2676
sll0406 (Protein) - TU2676
sll0408 (mRNA / CDS) - TU2672
sll0408 (Protein) - TU2672
sll0410 (mRNA / CDS) - TU2670
sll0410 (Protein) - TU2670
sll0412 (mRNA / CDS) - TU2670
sll0412 (Protein) - TU2670
sll0413 (mRNA / CDS) - TU2669
sll0413 (Protein) - TU2669
sll0413-5UTR (5'UTR) - TU2669
sll0414 (mRNA / CDS) - TU2665
sll0414 (Protein) - TU2665
sll0415 (mRNA / CDS) - TU2665
sll0415 (Protein) - TU2665
sll0418 (mRNA / CDS) - TU2181
sll0418 (Protein) - TU2181
sll0419 (mRNA / CDS) - TU2179
sll0419 (Protein) - TU2179
sll0422 (mRNA / CDS) - TU2176
sll0422 (Protein) - TU2176
sll0426 (mRNA / CDS) - TU2168
sll0426 (Protein) - TU2168
sll0426-5UTR (5'UTR) - TU2168
sll0428 (Protein)
sll0431 (transp mRNA / CDS)
sll0436 (mRNA / CDS)
sll0436 (Protein)
sll0441 (mRNA / CDS)
sll0441 (Protein)
sll0443 (mRNA / CDS)
sll0443 (Protein)
sll0444 (mRNA / CDS)
sll0444 (Protein)
sll0445 (mRNA / CDS)
sll0445 (Protein)
sll0446 (mRNA / CDS)
sll0446 (Protein)
sll0447 (mRNA / CDS)
sll0447 (Protein)
sll0451 (mRNA / CDS)
sll0451 (Protein)
sll0456 (mRNA / CDS) - TU2772
sll0456 (Protein) - TU2772
sll0456-5UTR (5'UTR) - TU2772
sll0462 (mRNA / CDS) - TU2744
sll0462 (Protein) - TU2744
sll0470 (mRNA / CDS) - TU3100
sll0470 (Protein) - TU3100
sll0471 (mRNA / CDS) - TU3100
sll0471 (Protein) - TU3100
sll0482 (mRNA / CDS) - TU3075
sll0482 (Protein) - TU3075
sll0485 (mRNA / CDS) - TU3068
sll0485 (Protein) - TU3068
sll0487 (mRNA / CDS) - TU3067
sll0487 (Protein) - TU3067
sll0488 (mRNA / CDS) - TU3065
sll0488 (Protein) - TU3065
sll0489 (mRNA / CDS) - TU3063
sll0489 (Protein) - TU3063
sll0493 (Protein) - TU3396
sll0497 (mRNA / CDS) - TU3391
sll0497 (Protein) - TU3391
sll0498 (mRNA / CDS) - TU3389
sll0498 (Protein) - TU3389
sll0498-3UTR (3'UTR) - TU3389
sll0499 (mRNA / CDS)
sll0499 (Protein)
sll0501 (mRNA / CDS) - TU3382
sll0501 (Protein) - TU3382
sll0505 (mRNA / CDS) - TU3377
sll0505 (Protein) - TU3377
sll0505-5UTR (5'UTR) - TU3377
sll0506 (mRNA / CDS) - TU3377
sll0506 (Protein) - TU3377
sll0508 (Protein)
sll0509 (mRNA / CDS) - TU3370
sll0509 (Protein) - TU3370
sll0513 (mRNA / CDS) - TU3354
sll0513 (Protein) - TU3354
sll0514 (mRNA / CDS) - TU3351
sll0514 (Protein) - TU3351
sll0518 (mRNA / CDS) - TU3342
sll0518 (Protein) - TU3342
sll0518-5UTR (5'UTR) - TU3342
sll0524 (mRNA / CDS) - TU3429
sll0524 (Protein) - TU3429
sll0525 (Protein) - TU3427
sll0528 (mRNA / CDS) - TU3422
sll0528 (Protein) - TU3422
sll0528-3UTR (3'UTR) - TU3422
sll0529 (mRNA / CDS) - TU3421
sll0529 (Protein) - TU3421
sll0529-3UTR (3'UTR) - TU3421
sll0540 (mRNA / CDS) - TU2979
sll0540 (Protein) - TU2979
sll0540-as (asRNA)
sll0544 (mRNA / CDS) - TU2959
sll0544 (Protein) - TU2959
sll0545 (mRNA / CDS) - TU2959
sll0545 (Protein) - TU2959
sll0546 (mRNA / CDS) - TU2958
sll0546 (Protein) - TU2958
sll0546-5UTR (5'UTR) - TU2958
sll0547 (mRNA / CDS) - TU2958
sll0547 (Protein) - TU2958
sll0553 (mRNA / CDS) - TU2950
sll0553 (Protein) - TU2950
sll0556 (mRNA / CDS) - TU2948
sll0556 (Protein) - TU2948
sll0563 (mRNA / CDS)
sll0563 (Protein)
sll0564 (Protein)
sll0565 (Protein) - TU3702
sll0572 (mRNA / CDS)
sll0572 (Protein)
sll0576 (mRNA / CDS)
sll0576 (Protein)
sll0577 (mRNA / CDS)
sll0577 (Protein)
sll0585 (mRNA / CDS) - TU3129
sll0585 (Protein) - TU3129
sll0586 (mRNA / CDS)
sll0586 (Protein)
sll0588 (mRNA / CDS) - TU3121
sll0588 (Protein) - TU3121
sll0588-3UTR (3'UTR) - TU3121
sll0590 (mRNA / CDS) - TU3115
sll0590 (Protein) - TU3115
sll0595 (mRNA / CDS) - TU3107
sll0595 (Protein) - TU3107
sll0596 (mRNA / CDS) - TU3106
sll0596 (Protein) - TU3106
sll0597 (mRNA / CDS) - TU3105
sll0597 (Protein) - TU3105
sll0601 (mRNA / CDS) - TU2826
sll0601 (Protein) - TU2826
sll0602 (mRNA / CDS) - TU2819
sll0602 (Protein) - TU2819
sll0602-5UTR (5'UTR) - TU2819
sll0606 (mRNA / CDS)
sll0606 (Protein)
sll0611 (mRNA / CDS) - TU2807
sll0611 (Protein) - TU2807
sll0611-3UTR (3'UTR) - TU2807
sll0614 (mRNA / CDS) - TU2803
sll0614 (Protein) - TU2803
sll0615 (mRNA / CDS) - TU2800
sll0615 (Protein) - TU2800
sll0623 (mRNA / CDS) - TU3527
sll0623 (Protein) - TU3527
sll0623-5UTR (5'UTR) - TU3527
sll0625 (mRNA / CDS) - TU3523
sll0625 (Protein) - TU3523
sll0630 (mRNA / CDS) - TU3501
sll0630 (Protein) - TU3501
sll0630-5UTR (5'UTR) - TU3501
sll0638 (mRNA / CDS) - TU427
sll0638 (Protein) - TU427
sll0639 (mRNA / CDS) - TU424
sll0639 (Protein) - TU424
sll0639-3UTR (3'UTR) - TU424
sll0639-5UTR (5'UTR) - TU424
sll0641 (mRNA / CDS)
sll0641 (Protein)
sll0644 (mRNA / CDS)
sll0644 (Protein)
sll0645 (mRNA / CDS) - TU406
sll0645 (Protein) - TU406
sll0647 (mRNA / CDS) - TU405
sll0647 (Protein) - TU405
sll0647-5UTR (5'UTR) - TU405
sll0649 (mRNA / CDS) - TU403
sll0649 (Protein) - TU403
sll0651 (transp mRNA / CDS) - TU402
sll0654 (mRNA / CDS) - TU388
sll0654 (Protein) - TU388
sll0654-as2 (asRNA)
sll0659 (mRNA / CDS) - TU3291
sll0659 (Protein) - TU3291
sll0662 (mRNA / CDS) - TU3286
sll0662 (Protein) - TU3286
sll0667 (transp mRNA / CDS) - TU3277
sll0670 (mRNA / CDS)
sll0670 (Protein)
sll0676 (Protein) - TU3263
sll0677 (transp mRNA / CDS) - TU3258
pstB/sll0683 (mRNA / CDS) - TU2785
sll0684,sll0683 (Protein)
sll0685 (mRNA / CDS) - TU2784
sll0685 (Protein) - TU2784
sll0685-5UTR (5'UTR) - TU2784
sll0691 (mRNA / CDS) - TU2777
sll0691 (Protein) - TU2777
sll0699 (transp mRNA / CDS) - TU112
sll0702 (mRNA / CDS)
sll0702 (Protein)
sll0703 (mRNA / CDS)
sll0703 (Protein)
sll0710 (mRNA / CDS) - TU97
sll0710 (Protein) - TU97
sll0723 (mRNA / CDS) - TU3613
sll0723 (Protein) - TU3613
sll0723-as (asRNA)
sll0729 (mRNA / CDS) - TU3600
sll0729 (Protein) - TU3600
sll0732 (Protein)
sll0732-as (asRNA)
sll0733 (mRNA / CDS) - TU3595
sll0733 (Protein) - TU3595
sll0733-5UTR (5'UTR) - TU3595
sll0735 (mRNA / CDS) - TU3590
sll0735 (Protein) - TU3590
sll0736 (mRNA / CDS) - TU3590
sll0736 (Protein) - TU3590
sll0737 (mRNA / CDS) - TU3300
sll0737 (Protein) - TU3300
sll0740 (mRNA / CDS) - TU3299
sll0740 (Protein) - TU3299
sll0744 (mRNA / CDS) - TU3296
sll0744 (Protein) - TU3296
sll0749 (mRNA / CDS) - TU2521
sll0749 (Protein) - TU2521
sll0752 (mRNA / CDS) - TU2516
sll0752 (Protein) - TU2516
sll0756 (mRNA / CDS) - TU2514
sll0756 (Protein) - TU2514
sll0759 (mRNA / CDS) - TU2510
sll0759 (Protein) - TU2510
sll0761 (mRNA / CDS) - TU2508
sll0761 (Protein) - TU2508
sll0762 (mRNA / CDS) - TU2508
sll0762 (Protein) - TU2508
sll0772 (mRNA / CDS)
sll0772 (Protein)
sll0777 (mRNA / CDS) - TU3253
sll0777 (Protein) - TU3253
sll0779 (mRNA / CDS)
sll0779 (Protein)
sll0781 (mRNA / CDS) - TU3247
sll0781 (Protein) - TU3247
sll0781-3UTR (3'UTR) - TU3247
sll0783 (Protein) - TU3241
sll0788 (mRNA / CDS) - TU3230
sll0788,slr6039 (Protein)
sll0788-5UTR (5'UTR) - TU3230
sll0789 (mRNA / CDS) - TU3230
sll0789,slr6040 (Protein)
hik31/sll0790 (mRNA / CDS) - TU3230
sll0800 (mRNA / CDS) - TU3210
sll0803 (mRNA / CDS) - TU1773
sll0803 (Protein) - TU1773
sll0804 (mRNA / CDS) - TU1773
sll0804 (Protein) - TU1773
sll0814 (mRNA / CDS) - TU1740
sll0814 (Protein) - TU1740
sll0814-5UTR (5'UTR) - TU1740
sll0815 (mRNA / CDS) - TU1737
sll0815 (Protein) - TU1737
sll0816 (mRNA / CDS) - TU1737
sll0816 (Protein) - TU1737
sll0822 (mRNA / CDS) - TU3017
sll0822 (Protein) - TU3017
sll0827 (mRNA / CDS) - TU3005
sll0827 (Protein) - TU3005
sll0833 (mRNA / CDS) - TU2991
sll0833 (Protein) - TU2991
sll0837 (mRNA / CDS) - TU1405
sll0837 (Protein) - TU1405
sll0839 (mRNA / CDS) - TU1405
sll0839 (Protein) - TU1405
sll0846 (mRNA / CDS) - TU1394
sll0846 (Protein) - TU1394
sll0846-3UTR (3'UTR) - TU1394
sll0846-5UTR (5'UTR) - TU1394
sll0854 (mRNA / CDS) - TU1381
sll0854 (Protein) - TU1381
sll0854-as (asRNA)
sll0855 (mRNA / CDS) - TU1381
sll0855 (Protein) - TU1381
sll0858 (mRNA / CDS) - TU1380
sll0858 (Protein) - TU1380
sll0860 (mRNA / CDS) - TU1200
sll0860 (Protein) - TU1200
sll0862 (mRNA / CDS) - TU1200
sll0862 (Protein) - TU1200
sll0867 (mRNA / CDS) - TU1193
sll0867 (Protein) - TU1193
sll0871 (mRNA / CDS) - TU1185
sll0871 (Protein) - TU1185
sll0872 (mRNA / CDS)
sll0872 (Protein)
sll0875 (mRNA / CDS) - TU1165
sll0875 (Protein) - TU1165
sll0877 (mRNA / CDS) - TU1161
sll0877 (Protein) - TU1161
sll0886 (mRNA / CDS) - TU2917
sll0886 (Protein) - TU2917
sll0887 (mRNA / CDS) - TU2916
sll0887 (Protein) - TU2916
sll0888 (Protein) - TU2915
sll0909 (Protein) - TU2090
sll0913 (mRNA / CDS) - TU2085
sll0913 (Protein) - TU2085
sll0914 (Protein)
sll0921 (mRNA / CDS) - TU2067
sll0921 (Protein) - TU2067
sll0925 (mRNA / CDS) - TU338
sll0925 (Protein) - TU338
sll0931 (mRNA / CDS) - TU333
sll0931 (Protein) - TU333
sll0933 (mRNA / CDS) - TU332
sll0933 (Protein) - TU332
sll0936 (mRNA / CDS) - TU327
sll0936 (Protein) - TU327
sll0938 (mRNA / CDS) - TU326
sll0938 (Protein) - TU326
sll0943 (mRNA / CDS) - TU316
sll0943 (Protein) - TU316
sll0943-3UTR (3'UTR) - TU316
sll0944 (mRNA / CDS) - TU2361
sll0944 (Protein) - TU2361
sll0944-as (asRNA)
sll0980 (mRNA / CDS) - TU665
sll0980 (Protein) - TU665
sll0980-5UTR (5'UTR) - TU665
sll0981 (mRNA / CDS) - TU665
sll0981 (Protein) - TU665
sll0982 (mRNA / CDS) - TU664
sll0982 (Protein) - TU664
sll0982-5UTR (5'UTR) - TU664
sll0985 (mRNA / CDS)
sll0985 (Protein)
sll0993 (mRNA / CDS) - TU633
sll0993 (Protein) - TU633
sll0994 (mRNA / CDS) - TU633
sll0994 (Protein) - TU633
sll0995 (mRNA / CDS) - TU633
sll0995 (Protein) - TU633
sll0997 (mRNA / CDS)
sll0997 (Protein)
sll1001 (mRNA / CDS) - TU504
sll1001 (Protein) - TU504
sll1004 (mRNA / CDS) - TU500
sll1004 (Protein) - TU500
sll1020 (mRNA / CDS) - TU378
sll1020 (Protein) - TU378
sll1021 (mRNA / CDS) - TU377
sll1021 (Protein) - TU377
sll1021-3UTR (3'UTR) - TU377
sll1021-5UTR (5'UTR) - TU377
sll1022 (mRNA / CDS) - TU374
sll1022 (Protein) - TU374
sll1022-5UTR (5'UTR) - TU374
sll1025 (mRNA / CDS) - TU371
sll1025 (Protein) - TU371
sll1033 (mRNA / CDS) - TU201
sll1033 (Protein) - TU201
sll1036 (mRNA / CDS) - TU198
sll1036 (Protein) - TU198
sll1039 (mRNA / CDS)
sll1039 (Protein)
sll1049 (mRNA / CDS) - TU83
sll1049 (Protein) - TU83
sll1053 (mRNA / CDS) - TU78
sll1053 (Protein) - TU78
sll1054 (mRNA / CDS) - TU78
sll1054 (Protein) - TU78
sll1060 (mRNA / CDS) - TU69
sll1060 (Protein) - TU69
sll1061 (mRNA / CDS) - TU67
sll1061 (Protein) - TU67
sll1061-5UTR (5'UTR) - TU67
sll1062 (Protein) - TU67
sll1063 (mRNA / CDS) - TU63
sll1063 (Protein) - TU63
sll1064 (mRNA / CDS)
sll1064 (Protein)
sll1071 (mRNA / CDS) - TU817
sll1071 (Protein) - TU817
sll1072 (mRNA / CDS) - TU817
sll1072 (Protein) - TU817
sll1080 (mRNA / CDS) - TU794
sll1080 (Protein) - TU794
sll1081 (mRNA / CDS) - TU794
sll1081 (Protein) - TU794
sll1084 (mRNA / CDS) - TU790
sll1084 (Protein) - TU790
sll1087 (mRNA / CDS) - TU2012
sll1087 (Protein) - TU2012
sll1087-5UTR (5'UTR) - TU2012
sll1089 (mRNA / CDS) - TU2012
sll1089 (Protein) - TU2012
sll1092 (Protein)
sll1106 (mRNA / CDS)
sll1106 (Protein)
sll1107 (mRNA / CDS)
sll1107 (Protein)
surE/sll1108 (mRNA / CDS)
sll1108 (Protein)
sll1109 (Protein)
sll1118 (Protein) - TU885
sll1120 (mRNA / CDS) - TU877
sll1120 (Protein) - TU877
sll1120-5UTR (5'UTR) - TU877
sll1121 (mRNA / CDS) - TU875
sll1121 (Protein) - TU875
sll1121-as3 (asRNA)
sll1123 (mRNA / CDS) - TU867
sll1123 (Protein) - TU867
sll1123-5UTR (5'UTR) - TU867
sll1123-as (asRNA)
sll1130 (mRNA / CDS) - TU1078
sll1130 (Protein) - TU1078
sll1130-as (asRNA)
sll1135 (mRNA / CDS) - TU1062
sll1135 (Protein) - TU1062
sll1138 (mRNA / CDS)
sll1138 (Protein)
sll1142 (mRNA / CDS) - TU1047
sll1142 (Protein) - TU1047
sll1147 (mRNA / CDS) - TU1441
sll1147 (Protein) - TU1441
sll1150 (mRNA / CDS) - TU1436
sll1150 (Protein) - TU1436
sll1151 (mRNA / CDS) - TU1431
sll1151 (Protein) - TU1431
sll1155 (mRNA / CDS) - TU1426
sll1155 (Protein) - TU1426
sll1155-5UTR (5'UTR) - TU1426
sll1156 (transp mRNA / CDS) - TU1426
sll1159 (mRNA / CDS) - TU1420
sll1159 (Protein) - TU1420
sll1160 (Protein) - TU1420
sll1162 (Protein) - TU1420
sll1163 (Protein) - TU1420
sll1166 (mRNA / CDS) - TU1962
sll1166 (Protein) - TU1962
sll1173 (Protein)
sll1174 (mRNA / CDS) - TU1949
sll1174 (Protein) - TU1949
sll1178 (mRNA / CDS) - TU1934
sll1178 (Protein) - TU1934
sll1186 (mRNA / CDS) - TU311
sll1186 (Protein) - TU311
sll1188 (mRNA / CDS) - TU310
sll1188 (Protein) - TU310
sll1193 (mRNA / CDS) - TU292
sll1193 (Protein) - TU292
sll1200 (mRNA / CDS) - TU284
sll1200 (Protein) - TU284
sll1201 (mRNA / CDS) - TU279
sll1201 (Protein) - TU279
sll1217 (mRNA / CDS) - TU1720
sll1217 (Protein) - TU1720
sll1222 (mRNA / CDS) - TU1714
sll1222 (Protein) - TU1714
sll1232 (Protein)
sll1233 (mRNA / CDS)
sll1233 (Protein)
sll1236 (Protein) - TU1694
sll1239 (mRNA / CDS) - TU1684
sll1239 (Protein) - TU1684
sll1241 (Protein) - TU1684
sll1242 (mRNA / CDS) - TU1811
sll1242 (Protein) - TU1811
sll1247 (mRNA / CDS) - TU1804
sll1247 (Protein) - TU1804
sll1247-as (asRNA)
sll1251 (mRNA / CDS) - TU1797
sll1251 (Protein) - TU1797
sll1252 (mRNA / CDS) - TU1794
sll1252 (Protein) - TU1794
sll1254 (mRNA / CDS) - TU1788
sll1254 (Protein) - TU1788
sll1255 (transp mRNA / CDS) - TU1788
sll1256 (transp mRNA / CDS) - TU1785
sll1263 (mRNA / CDS) - TU1776
sll1263 (Protein) - TU1776
sll1265 (mRNA / CDS) - TU1156
sll1265 (Protein) - TU1156
sll1265-as2 (asRNA)
sll1267 (mRNA / CDS) - TU1155
sll1267 (Protein) - TU1155
sll1267-3UTR (3'UTR) - TU1155
sll1268 (mRNA / CDS) - TU1153
sll1268 (Protein) - TU1153
sll1268-3UTR (3'UTR) - TU1153
sll1268-5UTR (5'UTR) - TU1153
sll1271 (mRNA / CDS) - TU1141
sll1271 (Protein) - TU1141
sll1272 (mRNA / CDS) - TU1141
sll1272 (Protein) - TU1141
sll1273 (mRNA / CDS) - TU1140
sll1273 (Protein) - TU1140
sll1274 (mRNA / CDS) - TU1135
sll1274 (Protein) - TU1135
sll1276 (mRNA / CDS) - TU1131
sll1276 (Protein) - TU1131
sll1280 (mRNA / CDS) - TU1125
sll1280 (Protein) - TU1125
sll1284 (mRNA / CDS) - TU702
sll1284 (Protein) - TU702
sll1286 (mRNA / CDS) - TU696
sll1286 (Protein) - TU696
sll1286-3UTR (3'UTR) - TU696
sll1289 (Protein) - TU683
sll1291 (mRNA / CDS) - TU681
sll1291 (Protein) - TU681
sll1292 (mRNA / CDS) - TU680
sll1292 (Protein) - TU680
sll1293 (mRNA / CDS) - TU680
sll1293 (Protein) - TU680
sll1304 (mRNA / CDS) - TU1224
sll1304 (Protein) - TU1224
sll1305 (mRNA / CDS) - TU1224
sll1305 (Protein) - TU1224
sll1306 (mRNA / CDS) - TU1224
sll1306 (Protein) - TU1224
sll1307 (mRNA / CDS) - TU1224
sll1307 (Protein) - TU1224
sll1308 (Protein) - TU1220
sll1315 (Protein)
sll1315-as (asRNA)
sll1318 (Protein) - TU178
sll1329 (mRNA / CDS) - TU3470
sll1329 (Protein) - TU3470
sll1330 (mRNA / CDS) - TU3469
sll1330 (Protein) - TU3469
sll1330-3UTR (3'UTR) - TU3469
sll1334 (mRNA / CDS)
sll1334 (Protein)
sll1336 (mRNA / CDS) - TU3457
sll1336 (Protein) - TU3457
sll1336-3UTR (3'UTR) - TU3457
sll1336-5UTR (5'UTR) - TU3457
sll1338 (mRNA / CDS) - TU3455
sll1338 (Protein) - TU3455
sll1338-3UTR (3'UTR) - TU3455
sll1338-5UTR (5'UTR) - TU3455
sll1350 (mRNA / CDS) - TU1122
sll1350 (Protein) - TU1122
sll1350-3UTR (3'UTR) - TU1122
sll1352 (mRNA / CDS) - TU1117
sll1352 (Protein) - TU1117
sll1355 (mRNA / CDS) - TU1105
sll1355 (Protein) - TU1105
sll1358 (mRNA / CDS) - TU1100
sll1358 (Protein) - TU1100
sll1359 (mRNA / CDS) - TU1099
sll1359 (Protein) - TU1099
sll1365 (mRNA / CDS) - TU3487
sll1365 (Protein) - TU3487
sll1366 (mRNA / CDS) - TU3484
sll1366 (Protein) - TU3484
sll1367 (mRNA / CDS) - TU3473
sll1367 (Protein) - TU3473
sll1369 (mRNA / CDS) - TU1926
sll1369 (Protein) - TU1926
sll1372 (mRNA / CDS) - TU1921
sll1372 (Protein) - TU1921
sll1376 (mRNA / CDS) - TU1915
sll1376 (Protein) - TU1915
sll1376-5UTR (5'UTR) - TU1915
sll1377 (mRNA / CDS) - TU1911
sll1377 (Protein) - TU1911
sll1380 (mRNA / CDS) - TU1906
sll1380 (Protein) - TU1906
sll1381 (mRNA / CDS) - TU1906
sll1381 (Protein) - TU1906
sll1383 (mRNA / CDS) - TU1903
sll1383 (Protein) - TU1903
sll1384 (mRNA / CDS) - TU1901
sll1384 (Protein) - TU1901
sll1384-3UTR (3'UTR) - TU1901
sll1384-5UTR (5'UTR) - TU1901
sll1386 (Protein) - TU1894
sll1388 (mRNA / CDS) - TU1889
sll1388 (Protein) - TU1889
sll1388-5UTR (5'UTR) - TU1889
sll1390 (mRNA / CDS) - TU1886
sll1390 (Protein) - TU1886
sll1396 (mRNA / CDS) - TU50
sll1396 (Protein) - TU50
sll1397 (transp mRNA / CDS) - TU50
sll1399 (mRNA / CDS) - TU46
sll1399 (Protein) - TU46
sll1400 (Protein) - TU46
sll1401 (mRNA / CDS) - TU46
sll1401 (Protein) - TU46
sll1414 (mRNA / CDS) - TU1643
sll1414 (Protein) - TU1643
sll1424 (mRNA / CDS) - TU1630
sll1424 (Protein) - TU1630
sll1426 (mRNA / CDS) - TU1624
sll1426 (Protein) - TU1624
sll1426-3UTR (3'UTR) - TU1624
sll1429 (mRNA / CDS) - TU1615
sll1429 (Protein) - TU1615
sll1436 (transp mRNA / CDS) - TU1971
sll1442 (Protein)
sll1446 (mRNA / CDS) - TU1962
sll1446 (Protein) - TU1962
sll1447 (Protein) - TU1962
sll1455 (Protein) - TU1022
sll1456 (Protein) - TU1018
surE/sll1459 (mRNA / CDS) - TU1013
sll1459 (Protein) - TU1013
sll1459-5UTR (5'UTR) - TU1013
sll1461 (mRNA / CDS) - TU1010
sll1461 (Protein) - TU1010
sll1464 (mRNA / CDS) - TU1008
sll1464 (Protein) - TU1008
sll1466 (mRNA / CDS) - TU1006
sll1466 (Protein) - TU1006
sll1469 (mRNA / CDS)
sll1469 (Protein)
sll1473 (mRNA / CDS) - TU3579
sll1473 (Protein) - TU3579
sll1474 (transp mRNA / CDS) - TU3579
sll1477 (mRNA / CDS) - TU3575
sll1477 (Protein) - TU3575
sll1481 (mRNA / CDS) - TU3572
sll1481 (Protein) - TU3572
sll1481-5UTR (5'UTR) - TU3572
sll1483 (mRNA / CDS) - TU3571
sll1483 (Protein) - TU3571
sll1486 (mRNA / CDS) - TU3570
sll1486 (Protein) - TU3570
sll1491 (mRNA / CDS) - TU3557
sll1491 (Protein) - TU3557
sll1491-3UTR (3'UTR) - TU3557
sll1495 (mRNA / CDS) - TU3546
sll1495 (Protein) - TU3544
sll1504 (Protein) - TU463
sll1505 (Protein) - TU463
sll1507 (mRNA / CDS) - TU457
sll1507 (Protein) - TU457
sll1509 (Protein) - TU450
sll1512 (mRNA / CDS) - TU447
sll1512 (Protein) - TU447
sll1516 (mRNA / CDS) - TU437
sll1516 (Protein) - TU437
sll1524 (mRNA / CDS)
sll1524 (Protein)
sll1524-as (asRNA)
sll1526 (mRNA / CDS) - TU2137
sll1526 (Protein) - TU2137
sll1527 (mRNA / CDS)
sll1527 (Protein)
sll1528 (mRNA / CDS) - TU2134
sll1528 (Protein) - TU2134
sll1528-5UTR (5'UTR) - TU2134
sll1530 (mRNA / CDS) - TU2134
sll1530 (Protein) - TU2134
sll1531 (mRNA / CDS) - TU2134
sll1531 (Protein) - TU2134
sll1532 (mRNA / CDS) - TU2128
sll1532 (Protein) - TU2128
sll1534 (mRNA / CDS) - TU2125
sll1534 (Protein) - TU2125
sll1537 (Protein) - TU2117
sll1541 (mRNA / CDS) - TU2107
sll1541 (Protein) - TU2107
sll1542 (mRNA / CDS)
sll1542 (Protein)
sll1549 (Protein) - TU3631
sll1558 (mRNA / CDS) - TU2047
sll1558 (Protein) - TU2047
sll1559 (mRNA / CDS) - TU2040
sll1559 (Protein) - TU2040
sll1560 (transp mRNA / CDS) - TU2040
sll1563 (mRNA / CDS) - TU2033
sll1563 (Protein) - TU2033
sll1568 (mRNA / CDS) - TU749
sll1568 (Protein) - TU749
sll1570 (mRNA / CDS)
sll1570 (Protein)
sll1570-as (asRNA)
sll1571 (mRNA / CDS) - TU742
sll1571 (Protein) - TU742
sll1571-5UTR (5'UTR) - TU742
sll1573 (mRNA / CDS) - TU730
sll1573 (Protein) - TU730
sll1582 (mRNA / CDS) - TU707
sll1582 (Protein) - TU707
sll1584 (Protein) - TU704
sll1592 (mRNA / CDS)
sll1592 (Protein)
sll1601 (mRNA / CDS)
sll1601 (Protein)
sll1613 (Protein) - TU1494
sll1618 (mRNA / CDS) - TU1364
sll1618 (Protein) - TU1364
sll1620 (mRNA / CDS) - TU1361
sll1620 (Protein) - TU1361
sll1621 (mRNA / CDS) - TU1359
sll1621 (Protein) - TU1359
sll1621-3UTR (3'UTR) - TU1359
sll1621-5UTR (5'UTR) - TU1359
sll1623 (mRNA / CDS) - TU1356
sll1623 (Protein) - TU1356
sll1624 (mRNA / CDS) - TU1355
sll1624 (Protein) - TU1355
sll1628 (mRNA / CDS) - TU1347
sll1628 (Protein) - TU1347
sll1634 (mRNA / CDS) - TU1026
sll1634 (Protein) - TU1026
sll1638 (mRNA / CDS) - TU524
sll1638 (Protein) - TU524
sll1638-3UTR (3'UTR) - TU524
sll1638-5UTR (5'UTR) - TU524
sll1640 (mRNA / CDS) - TU520
sll1640 (Protein) - TU520
sll1643 (mRNA / CDS) - TU515
sll1643 (Protein) - TU515
sll1643-3UTR (3'UTR) - TU515
sll1647 (mRNA / CDS) - TU509
sll1647 (Protein) - TU509
sll1651 (mRNA / CDS) - TU273
sll1651 (Protein) - TU273
sll1652 (mRNA / CDS) - TU273
sll1652 (Protein) - TU273
sll1654 (mRNA / CDS) - TU270
sll1654 (Protein) - TU270
sll1654-5UTR (5'UTR) - TU270
sll1656 (mRNA / CDS) - TU268
sll1656 (Protein) - TU268
sll1658 (Protein)
sll1660 (mRNA / CDS) - TU259
sll1660 (Protein) - TU259
sll1663 (mRNA / CDS) - TU253
sll1663 (Protein) - TU253
sll1664 (mRNA / CDS) - TU253
sll1664 (Protein) - TU253
sll1665 (mRNA / CDS) - TU251
sll1665 (Protein) - TU251
sll1665-3UTR (3'UTR) - TU251
sll1665-5UTR (5'UTR) - TU251
sll1665-as (asRNA)
sll1671 (Protein)
sll1673 (mRNA / CDS) - TU236
sll1673 (Protein) - TU236
sll1675 (mRNA / CDS) - TU233
sll1675 (Protein) - TU233
sll1677 (mRNA / CDS) - TU230
sll1677 (Protein) - TU230
sll1678 (Protein) - TU230
sll1680 (mRNA / CDS) - TU225
sll1680 (Protein) - TU225
sll1682 (mRNA / CDS) - TU223
sll1682 (Protein) - TU223
sll1682-5UTR (5'UTR) - TU223
sll1691 (mRNA / CDS)
sll1691 (Protein)
sll1692 (mRNA / CDS) - TU1335
sll1692 (Protein) - TU1335
sll1692-3UTR (3'UTR) - TU1335
sll1692-5UTR (5'UTR) - TU1335
sll1693 (mRNA / CDS) - TU1332
sll1693 (Protein) - TU1332
sll1693-3UTR (3'UTR) - TU1332
sll1696 (mRNA / CDS) - TU1330
sll1696 (Protein) - TU1330
sll1697 (mRNA / CDS) - TU1324
sll1697 (Protein) - TU1324
sll1697-3UTR (3'UTR) - TU1324
sll1698 (mRNA / CDS) - TU1322
sll1698 (Protein) - TU1322
sll1699 (mRNA / CDS) - TU1322
sll1699 (Protein) - TU1322
sll1703 (mRNA / CDS) - TU1318
sll1703 (Protein) - TU1318
sll1703-as (asRNA)
sll1708 (mRNA / CDS) - TU1312
sll1708 (Protein) - TU1312
sll1710 (transp mRNA / CDS) - TU1310
sll1712 (mRNA / CDS) - TU986
sll1712 (Protein) - TU986
sll1712-5UTR (5'UTR) - TU986
sll1714 (Protein) - TU984
sll1715 (Protein) - TU984
sll1716 (transp mRNA / CDS) - TU981
sll1723 (mRNA / CDS)
sll1723 (Protein)
sll1725 (mRNA / CDS)
sll1725 (Protein)
sll1725-as (asRNA)
sll1730 (mRNA / CDS) - TU954
sll1730 (Protein) - TU954
sll1730-as2 (asRNA)
sll1735 (mRNA / CDS) - TU949
sll1735 (Protein) - TU949
sll1735-3UTR (3'UTR) - TU949
sll1735-5UTR (5'UTR) - TU949
sll1739 (mRNA / CDS) - TU938
sll1739 (Protein) - TU938
sll1749 (mRNA / CDS) - TU537
sll1749 (Protein) - TU537
sll1757 (mRNA / CDS) - TU1271
sll1757 (Protein) - TU1271
sll1757-3UTR (3'UTR) - TU1271
sll1757-5UTR (5'UTR) - TU1271
sll1762 (mRNA / CDS) - TU1264
sll1762 (Protein) - TU1264
sll1764 (mRNA / CDS) - TU1262
sll1764 (Protein) - TU1262
sll1764-5UTR (5'UTR) - TU1262
sll1768 (mRNA / CDS) - TU1253
sll1768 (Protein) - TU1253
sll1769 (mRNA / CDS) - TU1249
sll1769 (Protein) - TU1249
sll1770 (mRNA / CDS) - TU1249
sll1770 (Protein) - TU1249
sll1773 (mRNA / CDS) - TU1246
sll1773 (Protein) - TU1246
sll1773-as (asRNA)
sll1775 (mRNA / CDS) - TU1237
sll1775 (Protein) - TU1237
sll1780 (transp mRNA / CDS) - TU1231
sll1783 (mRNA / CDS) - TU1224
sll1783 (Protein) - TU1224
sll1784 (mRNA / CDS) - TU1224
sll1784 (Protein) - TU1224
sll1785 (mRNA / CDS) - TU1224
sll1785 (Protein) - TU1224
sll1791 (transp mRNA / CDS) - TU863
sll1825 (mRNA / CDS) - TU828
sll1825 (Protein) - TU828
sll1825-3UTR (3'UTR) - TU828
sll1825-as (asRNA)
sll1830 (mRNA / CDS) - TU621
sll1830 (Protein) - TU621
sll1834 (mRNA / CDS) - TU608
sll1834 (Protein) - TU608
sll1835 (mRNA / CDS) - TU606
sll1835 (Protein) - TU606
sll1835-3UTR (3'UTR) - TU606
sll1837 (mRNA / CDS) - TU602
sll1837 (Protein) - TU602
sll1837-3UTR (3'UTR) - TU602
sll1837-5UTR (5'UTR) - TU602
sll1848 (mRNA / CDS) - TU2322
sll1848 (Protein) - TU2322
sll1862 (mRNA / CDS) - TU2304
sll1862 (Protein) - TU2304
sll1862-3UTR (3'UTR) - TU2304
sll1863 (mRNA / CDS) - TU2303
sll1863 (Protein) - TU2303
sll1863-5UTR (5'UTR) - TU2303
sll1866 (mRNA / CDS) - TU1886
sll1866 (Protein) - TU1886
sll1870 (mRNA / CDS) - TU1876
sll1870 (Protein) - TU1876
sll1873 (mRNA / CDS) - TU1872
sll1873 (Protein) - TU1872
sll1873-3UTR (3'UTR) - TU1872
sll1878 (mRNA / CDS) - TU1867
sll1878 (Protein) - TU1867
sll1878-5UTR (5'UTR) - TU1867
sll1880 (mRNA / CDS) - TU1865
sll1880 (Protein) - TU1865
sll1880-3UTR (3'UTR) - TU1865
sll1882 (mRNA / CDS) - TU1862
sll1882 (Protein) - TU1862
sll1882-5UTR (5'UTR) - TU1862
sll1884 (mRNA / CDS) - TU1859
sll1884 (Protein) - TU1859
sll1885 (mRNA / CDS) - TU1854
sll1885 (Protein) - TU1854
sll1886 (mRNA / CDS) - TU1853
sll1886 (Protein) - TU1853
sll1891 (mRNA / CDS) - TU1834
sll1891 (Protein) - TU1834
sll1891-3UTR (3'UTR) - TU1834
sll1891-5UTR (5'UTR) - TU1834
sll1892 (mRNA / CDS) - TU1832
sll1892 (Protein) - TU1832
sll1895 (mRNA / CDS) - TU1825
sll1895 (Protein) - TU1825
sll1897 (mRNA / CDS) - TU1821
sll1897 (Protein) - TU1821
sll1897-5UTR (5'UTR) - TU1821
sll1902 (mRNA / CDS) - TU1489
sll1902 (Protein) - TU1489
sll1906 (mRNA / CDS) - TU1479
sll1906 (Protein) - TU1479
sll1906-as2 (asRNA)
sll1911 (Protein) - TU783
sll1911-3UTR (3'UTR) - TU783
sll1913 (mRNA / CDS) - TU780
sll1913 (Protein) - TU780
sll1915 (mRNA / CDS) - TU774
sll1915 (Protein) - TU774
sll1915-5UTR (5'UTR) - TU774
sll1921 (mRNA / CDS) - TU766
sll1921 (Protein) - TU766
sll1925 (mRNA / CDS) - TU755
sll1925 (Protein) - TU755
sll1925-3UTR (3'UTR) - TU755
sll1926 (mRNA / CDS) - TU752
sll1926 (Protein) - TU752
sll1926-3UTR (3'UTR) - TU752
sll1927 (mRNA / CDS) - TU750
sll1927 (Protein) - TU750
sll1927-5UTR (5'UTR) - TU750
sll1930 (transp mRNA / CDS) - TU593
sll1938 (mRNA / CDS) - TU568
sll1938 (Protein) - TU568
sll1939 (mRNA / CDS) - TU564
sll1939 (Protein) - TU564
sll1940 (mRNA / CDS) - TU564
sll1940 (Protein) - TU564
sll1942 (mRNA / CDS) - TU556
sll1942 (Protein) - TU556
sll1946 (mRNA / CDS)
sll1946 (Protein)
sll1949 (mRNA / CDS) - TU1469
sll1949 (Protein) - TU1469
sll1949-3UTR (3'UTR) - TU1469
sll1949-5UTR (5'UTR) - TU1469
sll1950 (mRNA / CDS) - TU1467
sll1950 (Protein) - TU1467
sll1950-3UTR (3'UTR) - TU1467
sll1956 (Protein) - TU1458
sll1959 (Protein) - TU1451
sll1961 (mRNA / CDS) - TU1449
sll1961 (Protein) - TU1449
sll1961-3UTR (3'UTR) - TU1449
sll1967 (mRNA / CDS) - TU915
sll1967 (Protein) - TU915
sll1971 (mRNA / CDS) - TU909
sll1971 (Protein) - TU909
sll1979 (mRNA / CDS) - TU1598
sll1979 (Protein) - TU1598
sll1982 (transp mRNA / CDS) - TU1590
sll1983 (transp mRNA / CDS) - TU1589
sll1998 (transp mRNA / CDS) - TU1669
sll2002 (mRNA / CDS) - TU1307
sll2002 (Protein) - TU1307
sll2002-3UTR (3'UTR) - TU1307
sll2003 (mRNA / CDS) - TU1304
sll2003 (Protein) - TU1304
sll2006 (mRNA / CDS) - TU1298
sll2006 (Protein) - TU1298
sll2006-as (asRNA)
sll2008 (mRNA / CDS) - TU1295
sll2008 (Protein) - TU1295
sll2009 (mRNA / CDS) - TU1295
sll2009 (Protein) - TU1295
sll2015 (mRNA / CDS) - TU1286
sll2015 (Protein) - TU1286
sll5004 (Protein) - TU5003
sll5033 (mRNA / CDS) - TU5036
sll5033 (Protein) - TU5036
sll5034 (mRNA / CDS) - TU5036
sll5034 (Protein) - TU5036
sll5035 (mRNA / CDS) - TU5036
sll5035 (Protein) - TU5036
sll5042 (Protein) - TU5047
sll5048 (mRNA / CDS) - TU5054
sll5048 (Protein) - TU5054
sll5048-5UTR (5'UTR) - TU5054
sll5050 (mRNA / CDS)
sll5050 (Protein)
sll5052 (mRNA / CDS) - TU5058
sll5052 (Protein) - TU5058
sll5059 (mRNA / CDS) - TU5069
sll5059 (Protein) - TU5069
sll5060 (mRNA / CDS) - TU5073
sll5060 (Protein) - TU5073
sll5061 (mRNA / CDS) - TU5073
sll5061 (Protein) - TU5073
sll5066 (Protein)
sll5075 (Protein) - TU5088
sll5079 (mRNA / CDS) - TU5092
sll5079 (Protein) - TU5092
sll5080 (mRNA / CDS) - TU5092
sll5080 (Protein) - TU5092
sll5081 (mRNA / CDS)
sll5081 (Protein)
sll5084 (mRNA / CDS)
sll5084,slr6070,slr6011 (Protein)
sll5094 (Protein)
sll5128 (mRNA / CDS) - TU5129
sll5128 (Protein) - TU5129
sll5131 (transp mRNA / CDS)
sll6017 (mRNA / CDS) - TU6016
sll6017-3UTR (3'UTR) - TU6016
sll6054 (mRNA / CDS) - TU6049
sll6054 (Protein) - TU6049
sll6055 (mRNA / CDS) - TU6049
sll6055 (Protein) - TU6049
sll6060 (mRNA / CDS)
sll6060 (Protein)
sll6076 (mRNA / CDS) - TU6071
sll6076,sll6017,sll0800 (Protein)
sll6076-3UTR (3'UTR) - TU6071
sll6093 (mRNA / CDS) - TU6086
sll6093 (Protein) - TU6086
sll7002 (transp mRNA / CDS)
sll7003 (mRNA / CDS) - TU7001
sll7003 (Protein) - TU7001
sll7006 (mRNA / CDS) - TU7002
sll7006 (Protein) - TU7002
sll7033 (Protein) - TU7026
sll7034 (mRNA / CDS) - TU7026
sll7034 (Protein) - TU7026
sll7043 (Protein) - TU7035
sll7044 (mRNA / CDS) - TU7035
sll7044 (Protein) - TU7035
sll7062 (mRNA / CDS) - TU7058
sll7062 (Protein) - TU7058
sll7069 (Protein) - TU7065
sll7070 (Protein) - TU7065
sll7078 (mRNA / CDS)
sll7078 (Protein)
sll7085 (mRNA / CDS) - TU7078
sll7085 (Protein) - TU7078
sll7086 (mRNA / CDS) - TU7078
sll7086 (Protein) - TU7078
sll7087 (mRNA / CDS) - TU7078
sll7087 (Protein) - TU7078
sll7089 (mRNA / CDS) - TU7079
sll7089 (Protein) - TU7079
sll7090 (mRNA / CDS) - TU7079
sll7090 (Protein) - TU7079
sll7103 (Protein)
sll8001 (mRNA / CDS) - TU8002
sll8001 (Protein) - TU8002
sll8002 (mRNA / CDS) - TU8005
sll8002 (Protein) - TU8005
sll8004 (mRNA / CDS) - TU8005
sll8004 (Protein) - TU8005
sll8007 (mRNA / CDS) - TU8005
sll8007 (Protein) - TU8005
sll8007-5UTR (5'UTR) - TU8005
sll8011 (mRNA / CDS) - TU8008
sll8011 (Protein) - TU8008
sll8012 (Protein) - TU8008
sll8027 (Protein) - TU8024
sll8043 (transp mRNA / CDS) - TU8041
sll8049 (mRNA / CDS)
sll8049 (Protein)
slr0001 (mRNA / CDS) - TU2568
slr0001 (Protein) - TU2568
slr0001-3UTR (3'UTR) - TU2568
slr0006 (mRNA / CDS) - TU2578
slr0006 (Protein) - TU2578
slr0006-5UTR (5'UTR) - TU2578
slr0007 (mRNA / CDS) - TU2578
slr0007 (Protein) - TU2578
slr0013 (mRNA / CDS) - TU2587
slr0013 (Protein) - TU2587
sppA/slr0021 (mRNA / CDS) - TU2608
slr0021 (Protein) - TU2608
slr0021-3UTR (3'UTR) - TU2608
slr0022 (mRNA / CDS) - TU3306
slr0022 (Protein) - TU3306
slr0031 (mRNA / CDS) - TU3316
slr0031 (Protein) - TU3316
slr0038 (mRNA / CDS) - TU3330
slr0038 (Protein) - TU3330
slr0039 (mRNA / CDS) - TU3330
slr0039 (Protein) - TU3330
slr0042 (mRNA / CDS) - TU3332
slr0042 (Protein) - TU3332
slr0042-as (asRNA)
slr0042-as2 (asRNA)
slr0049 (mRNA / CDS) - TU2696
slr0049 (Protein) - TU2696
slr0058 (mRNA / CDS)
slr0058 (Protein)
slr0060 (mRNA / CDS) - TU2718
slr0060 (Protein) - TU2718
slr0064 (mRNA / CDS) - TU2725
slr0064 (Protein) - TU2725
slr0065 (Protein) - TU2729
slr0069 (Protein)
slr0076 (mRNA / CDS) - TU3030
slr0076 (Protein) - TU3030
slr0086 (mRNA / CDS) - TU3053
slr0086 (Protein) - TU3053
slr0099 (transp mRNA / CDS) - TU3135
slr0105 (mRNA / CDS) - TU3149
slr0105 (Protein) - TU3149
slr0106 (mRNA / CDS) - TU3149
slr0106 (Protein) - TU3149
slr0108 (mRNA / CDS) - TU3152
slr0108 (Protein) - TU3152
slr0110 (Protein) - TU3155
slr0111 (mRNA / CDS) - TU3155
slr0111 (Protein) - TU3155
slr0112 (mRNA / CDS) - TU3159
slr0112 (Protein) - TU3159
slr0112-5UTR (5'UTR) - TU3159
slr0114 (mRNA / CDS) - TU3160
slr0114 (Protein) - TU3160
trmJ/slr0120 (mRNA / CDS) - TU3167
slr0120 (Protein) - TU3167
slr0121 (mRNA / CDS) - TU3168
slr0121 (Protein) - TU3168
slr0121-5UTR (5'UTR) - TU3168
slr0144 (mRNA / CDS) - TU2264
slr0144 (Protein) - TU2264
slr0144-5UTR (5'UTR) - TU2264
slr0145 (mRNA / CDS) - TU2264
slr0145 (Protein) - TU2264
slr0146 (mRNA / CDS) - TU2264
slr0146 (Protein) - TU2264
slr0147 (mRNA / CDS) - TU2264
slr0147 (Protein) - TU2264
slr0148 (mRNA / CDS) - TU2264
slr0148 (Protein) - TU2264
slr0149 (mRNA / CDS) - TU2264
slr0149 (Protein) - TU2264
slr0151 (mRNA / CDS) - TU2264
slr0151 (Protein) - TU2264
slr0168 (mRNA / CDS) - TU2391
slr0168 (Protein) - TU2391
slr0169 (mRNA / CDS) - TU2393
slr0169 (Protein) - TU2393
slr0180 (transp mRNA / CDS) - TU2414
slr0184 (mRNA / CDS) - TU2428
slr0184 (Protein) - TU2428
slr0192 (mRNA / CDS) - TU2876
slr0192 (Protein) - TU2876
slr0192-as (asRNA)
slr0196 (Protein) - TU2881
slr0199 (Protein) - TU2885
slr0207 (mRNA / CDS) - TU2902
slr0207 (Protein) - TU2902
slr0208 (mRNA / CDS) - TU2904
slr0208 (Protein) - TU2904
slr0208-as (asRNA)
slr0209 (mRNA / CDS) - TU2907
slr0209 (Protein) - TU2907
slr0211 (mRNA / CDS) - TU2911
slr0211 (Protein) - TU2911
slr0211-5UTR (5'UTR) - TU2911
slr0217 (mRNA / CDS) - TU2622
slr0217 (Protein) - TU2622
slr0226 (mRNA / CDS) - TU2648
slr0226 (Protein) - TU2648
slr0226-3UTR (3'UTR) - TU2648
slr0226-5UTR (5'UTR) - TU2648
slr0230 (transp mRNA / CDS) - TU2659
slr0238 (Protein) - TU122
slr0243 (mRNA / CDS) - TU127
slr0243 (Protein) - TU127
slr0244 (mRNA / CDS) - TU130
slr0244 (Protein) - TU130
slr0244-3UTR (3'UTR) - TU130
slr0244-5UTR (5'UTR) - TU130
slr0245 (mRNA / CDS) - TU135
slr0245 (Protein) - TU135
slr0245-5UTR (5'UTR) - TU135
slr0249 (mRNA / CDS) - TU139
slr0249 (Protein) - TU139
slr0250 (mRNA / CDS) - TU141
slr0250 (Protein) - TU141
slr0250-5UTR (5'UTR) - TU141
slr0254 (mRNA / CDS) - TU151
slr0254 (Protein) - TU151
slr0262 (mRNA / CDS) - TU1546
slr0262 (Protein) - TU1546
slr0263 (mRNA / CDS) - TU1546
slr0263 (Protein) - TU1546
slr0265 (transp mRNA / CDS) - TU1549
slr0267 (mRNA / CDS)
slr0267 (Protein)
slr0267-as (asRNA)
slr0269 (mRNA / CDS)
slr0269 (Protein)
slr0270 (mRNA / CDS) - TU1563
slr0270 (Protein) - TU1563
slr0270-3UTR (3'UTR) - TU1563
slr0280 (mRNA / CDS) - TU2189
slr0280 (Protein) - TU2189
slr0287 (mRNA / CDS) - TU2205
slr0287 (Protein) - TU2205
slr0291 (Protein) - TU2207
slr0300 (Protein)
slr0300-as (asRNA)
slr0302 (mRNA / CDS) - TU3182
slr0302 (Protein) - TU3182
slr0303 (Protein) - TU3187
slr0306 (mRNA / CDS) - TU3189
slr0306 (Protein) - TU3189
slr0309 (mRNA / CDS) - TU3196
slr0309 (Protein) - TU3196
slr0312 (mRNA / CDS) - TU3200
slr0312 (Protein) - TU3200
slr0315 (mRNA / CDS)
slr0315 (Protein)
slr0317 (mRNA / CDS)
slr0317 (Protein)
slr0318 (mRNA / CDS)
slr0318 (Protein)
slr0320 (mRNA / CDS) - TU2363
slr0320 (Protein) - TU2363
slr0320-3UTR (3'UTR) - TU2363
slr0320-as (asRNA)
slr0320-as2 (asRNA)
slr0320-as3 (asRNA)
slr0320-as4 (asRNA)
slr0325 (Protein) - TU2368
slr0326 (mRNA / CDS) - TU2370
slr0326 (Protein) - TU2370
slr0326-5UTR (5'UTR) - TU2370
slr0328 (Protein) - TU2370
slr0333 (mRNA / CDS) - TU2380
slr0333 (Protein) - TU2380
slr0334 (mRNA / CDS) - TU2380
slr0334 (Protein) - TU2380
slr0337 (mRNA / CDS) - TU2387
slr0337 (Protein) - TU2387
slr0338 (mRNA / CDS) - TU2391
slr0338 (Protein) - TU2391
slr0341 (mRNA / CDS)
slr0341 (Protein)
slr0345 (mRNA / CDS) - TU2530
slr0345 (Protein) - TU2530
slr0350 (transp mRNA / CDS)
slr0351 (mRNA / CDS) - TU2543
slr0351 (Protein) - TU2543
slr0351-as (asRNA)
slr0352 (transp mRNA / CDS) - TU2545
slr0354 (Protein) - TU2549
slr0355 (mRNA / CDS) - TU2555
slr0355 (Protein) - TU2555
slr0356 (mRNA / CDS) - TU2555
slr0356 (Protein) - TU2555
slr0358 (mRNA / CDS) - TU2561
slr0358 (Protein) - TU2561
slr0358-as (asRNA)
slr0359 (mRNA / CDS) - TU2432
slr0359 (Protein) - TU2432
slr0361 (mRNA / CDS) - TU2437
slr0361 (Protein) - TU2437
slr0362 (mRNA / CDS) - TU2438
slr0362 (Protein) - TU2438
slr0369 (mRNA / CDS) - TU2453
slr0369 (Protein) - TU2453
slr0373 (mRNA / CDS) - TU2462
slr0373 (Protein) - TU2462
slr0373-5UTR (5'UTR) - TU2462
slr0374 (mRNA / CDS) - TU2462
slr0374 (Protein) - TU2462
slr0376 (mRNA / CDS) - TU2463
slr0376 (Protein) - TU2463
slr0376-5UTR (5'UTR) - TU2463
slr0377 (mRNA / CDS) - TU2464
slr0377 (Protein) - TU2464
slr0378 (mRNA / CDS) - TU2466
slr0378 (Protein) - TU2466
slr0379 (mRNA / CDS) - TU2466
slr0379 (Protein) - TU2466
slr0380 (mRNA / CDS) - TU2469
slr0380 (Protein) - TU2469
slr0386 (mRNA / CDS) - TU2484
slr0386 (Protein) - TU2484
slr0397 (mRNA / CDS) - TU2224
slr0397 (Protein) - TU2224
slr0398 (mRNA / CDS) - TU2226
slr0398 (Protein) - TU2226
slr0401 (mRNA / CDS) - TU2228
slr0401 (Protein) - TU2228
slr0402 (mRNA / CDS) - TU2228
slr0402 (Protein) - TU2228
slr0404 (mRNA / CDS) - TU2233
slr0404 (Protein) - TU2233
slr0404-3UTR (3'UTR) - TU2233
slr0404-5UTR (5'UTR) - TU2233
slr0407 (mRNA / CDS)
slr0407 (Protein)
slr0408 (mRNA / CDS) - TU2243
slr0408 (Protein) - TU2243
slr0408-5UTR (5'UTR) - TU2243
slr0416 (mRNA / CDS) - TU2835
slr0416,sll1630 (Protein)
slr0416-3UTR (3'UTR) - TU2835
slr0418 (mRNA / CDS) - TU2836
slr0418 (Protein) - TU2836
slr0420 (mRNA / CDS) - TU2849
slr0420 (Protein) - TU2849
slr0427 (mRNA / CDS) - TU2873
slr0427 (Protein) - TU2873
slr0431 (mRNA / CDS) - TU2668
slr0431 (Protein) - TU2668
slr0431-3UTR (3'UTR) - TU2668
slr0439 (mRNA / CDS) - TU2684
slr0439 (Protein) - TU2684
slr0440 (mRNA / CDS) - TU2695
slr0440 (Protein) - TU2695
slr0440-5UTR (5'UTR) - TU2695
slr0442 (mRNA / CDS) - TU2158
slr0442 (Protein) - TU2158
slr0442-3UTR (3'UTR) - TU2158
slr0442-5UTR (5'UTR) - TU2158
slr0442-as (asRNA)
slr0442-as2 (asRNA)
slr0443 (mRNA / CDS) - TU2169
slr0443 (Protein) - TU2169
slr0443-3UTR (3'UTR) - TU2169
slr0445 (mRNA / CDS) - TU2178
slr0445 (Protein) - TU2178
slr0453 (mRNA / CDS) - TU3672
slr0453 (Protein) - TU3672
slr0454 (mRNA / CDS) - TU3673
slr0454 (Protein) - TU3673
slr0455 (mRNA / CDS) - TU3675
slr0455 (Protein) - TU3675
slr0458 (Protein)
slr0459 (mRNA / CDS) - TU3676
slr0459 (Protein) - TU3676
slr0460 (transp mRNA / CDS) - TU3677
slr0476 (mRNA / CDS) - TU2753
slr0476 (Protein) - TU2753
slr0476-3UTR (3'UTR) - TU2753
slr0476-5UTR (5'UTR) - TU2753
slr0479 (mRNA / CDS) - TU2755
slr0479 (Protein) - TU2755
slr0479-as (asRNA)
slr0479-as2 (asRNA)
slr0482 (mRNA / CDS) - TU2755
slr0482 (Protein) - TU2755
slr0483 (mRNA / CDS) - TU2760
slr0483 (Protein) - TU2760
slr0483-3UTR (3'UTR) - TU2760
slr0483-5UTR (5'UTR) - TU2760
slr0483-as (asRNA)
slr0487 (mRNA / CDS) - TU2766
slr0487 (Protein) - TU2766
slr0489 (mRNA / CDS) - TU2770
slr0489 (Protein) - TU2770
slr0491 (mRNA / CDS) - TU2770
slr0491 (Protein) - TU2770
slr0510 (mRNA / CDS)
slr0510 (Protein)
slr0511 (transp mRNA / CDS)
slr0514 (Protein)
slr0516 (mRNA / CDS) - TU3089
slr0516 (Protein) - TU3089
slr0535 (mRNA / CDS) - TU3363
slr0535 (Protein) - TU3363
slr0535-5UTR (5'UTR) - TU3363
slr0537 (mRNA / CDS) - TU3371
slr0537 (Protein) - TU3371
slr0541 (Protein) - TU3383
slr0544 (mRNA / CDS) - TU3388
slr0544 (Protein) - TU3388
slr0545 (mRNA / CDS) - TU3392
slr0545 (Protein) - TU3392
slr0552 (mRNA / CDS) - TU3419
slr0552 (Protein) - TU3419
slr0556 (mRNA / CDS) - TU3424
slr0556 (Protein) - TU3424
slr0565 (mRNA / CDS) - TU2953
slr0565 (Protein) - TU2953
slr0565-3UTR (3'UTR) - TU2953
slr0565-5UTR (5'UTR) - TU2953
slr0569 (mRNA / CDS) - TU2957
slr0569 (Protein) - TU2957
slr0569-3UTR (3'UTR) - TU2957
slr0573 (Protein) - TU2963
slr0575 (mRNA / CDS) - TU2967
slr0575 (Protein) - TU2967
slr0580 (mRNA / CDS) - TU2972
slr0580 (Protein) - TU2972
slr0581 (mRNA / CDS) - TU2976
slr0581 (Protein) - TU2976
slr0581-5UTR (5'UTR) - TU2976
slr0582 (mRNA / CDS) - TU2976
slr0582 (Protein) - TU2976
slr0583 (mRNA / CDS) - TU2982
slr0583 (Protein) - TU2982
slr0586 (Protein) - TU3689
slr0588 (mRNA / CDS)
slr0588 (Protein)
slr0589 (mRNA / CDS) - TU3691
slr0589 (Protein) - TU3691
slr0589-5UTR (5'UTR) - TU3691
slr0590 (mRNA / CDS) - TU3691
slr0590 (Protein) - TU3691
slr0591 (mRNA / CDS) - TU3694
slr0591 (Protein) - TU3694
slr0592 (Protein) - TU3697
slr0594 (mRNA / CDS) - TU3699
slr0594 (Protein) - TU3699
slr0594-3UTR (3'UTR) - TU3699
slr0596 (mRNA / CDS) - TU3701
slr0596 (Protein) - TU3701
slr0596-5UTR (5'UTR) - TU3701
slr0598 (mRNA / CDS) - TU3703
slr0598 (Protein) - TU3703
slr0598-5UTR (5'UTR) - TU3703
slr0599 (mRNA / CDS) - TU3704
slr0599 (Protein) - TU3704
slr0599-as (asRNA)
slr0600 (mRNA / CDS) - TU3706
slr0600 (Protein) - TU3706
slr0600-5UTR (5'UTR) - TU3706
slr0601 (mRNA / CDS) - TU3707
slr0601 (Protein) - TU3707
slr0602 (mRNA / CDS) - TU3708
slr0602 (Protein) - TU3708
slr0602-5UTR (5'UTR) - TU3708
slr0605 (mRNA / CDS) - TU3713
slr0605 (Protein) - TU3713
slr0606 (mRNA / CDS) - TU3714
slr0606 (Protein) - TU3714
slr0607 (Protein) - TU3715
slr0609 (mRNA / CDS) - TU3717
slr0609 (Protein) - TU3717
slr0613 (mRNA / CDS) - TU4
slr0613 (Protein) - TU4
slr0615 (mRNA / CDS) - TU3110
slr0615 (Protein) - TU3110
slr0615-as (asRNA)
slr0617 (mRNA / CDS) - TU3114
slr0617 (Protein) - TU3114
slr0619 (mRNA / CDS) - TU3119
slr0619 (Protein) - TU3119
slr0619-as (asRNA)
slr0624 (mRNA / CDS) - TU3132
slr0624 (Protein) - TU3132
slr0625 (mRNA / CDS) - TU3133
slr0625 (Protein) - TU3133
slr0625-5UTR (5'UTR) - TU3133
slr0626 (mRNA / CDS) - TU3135
slr0626 (Protein) - TU3135
slr0630 (mRNA / CDS) - TU2796
slr0630 (Protein) - TU2796
slr0630-5UTR (5'UTR) - TU2796
slr0634 (mRNA / CDS) - TU2806
slr0634 (Protein) - TU2806
slr0635 (mRNA / CDS) - TU2806
slr0635 (Protein) - TU2806
slr0637 (mRNA / CDS) - TU2811
slr0637 (Protein) - TU2811
slr0643 (mRNA / CDS) - TU2816
slr0643 (Protein) - TU2816
slr0643-3UTR (3'UTR) - TU2816
slr0643-5UTR (5'UTR) - TU2816
slr0645 (mRNA / CDS) - TU2821
slr0645 (Protein) - TU2821
slr0645-as2 (asRNA)
slr0650 (mRNA / CDS) - TU2828
slr0650 (Protein) - TU2828
slr0651 (mRNA / CDS) - TU3499
slr0651 (Protein) - TU3499
slr0654 (mRNA / CDS) - TU3510
slr0654 (Protein) - TU3510
slr0658 (mRNA / CDS) - TU3516
slr0658 (Protein) - TU3516
slr0662 (mRNA / CDS) - TU3525
slr0662 (Protein) - TU3525
slr0664 (Protein) - TU3529
slr0666 (mRNA / CDS) - TU3530
slr0666 (Protein) - TU3530
slr0667 (mRNA / CDS) - TU3532
slr0667 (Protein) - TU3532
slr0670 (mRNA / CDS) - TU386
slr0670 (Protein) - TU386
slr0678 (mRNA / CDS) - TU393
slr0678 (Protein) - TU393
slr0680 (mRNA / CDS) - TU399
slr0680 (Protein) - TU399
slr0680-5UTR (5'UTR) - TU399
slr0681 (mRNA / CDS) - TU399
slr0681 (Protein) - TU399
slr0686 (mRNA / CDS)
slr0686 (Protein)
slr0688 (mRNA / CDS)
slr0688 (Protein)
slr0688-as (asRNA)
slr0689 (mRNA / CDS) - TU422
slr0689 (Protein) - TU422
slr0695 (mRNA / CDS) - TU430
slr0695 (Protein) - TU430
slr0697 (mRNA / CDS)
slr0697 (Protein)
slr0698 (Protein)
slr0699 (mRNA / CDS)
slr0699 (Protein)
slr0703 (transp mRNA / CDS) - TU3265
slr0704 (transp mRNA / CDS) - TU3265
slr0708 (mRNA / CDS) - TU3274
slr0708 (Protein) - TU3274
slr0709 (mRNA / CDS) - TU3282
slr0709 (Protein) - TU3282
slr0719 (mRNA / CDS)
slr0719 (Protein)
slr0723 (mRNA / CDS) - TU103
slr0723 (Protein) - TU103
slr0723-5UTR (5'UTR) - TU103
slr0727 (mRNA / CDS) - TU104
slr0727 (Protein) - TU104
slr0728 (mRNA / CDS) - TU105
slr0728 (Protein) - TU105
slr0729 (mRNA / CDS) - TU107
slr0729 (Protein) - TU107
slr0729-5UTR (5'UTR) - TU107
slr0730 (mRNA / CDS) - TU107
slr0730 (Protein) - TU107
slr0731 (mRNA / CDS) - TU107
slr0731 (Protein) - TU107
slr0734 (mRNA / CDS) - TU111
slr0734 (Protein) - TU111
slr0734-3UTR (3'UTR) - TU111
slr0740 (mRNA / CDS) - TU116
slr0740 (Protein) - TU116
slr0751 (mRNA / CDS) - TU3596
slr0751 (Protein) - TU3596
slr0755 (Protein) - TU3608
slr0765 (mRNA / CDS) - TU3294
slr0765 (Protein) - TU3294
slr0769 (mRNA / CDS) - TU2489
slr0769 (Protein) - TU2489
slr0770 (Protein) - TU2489
slr0771 (Protein) - TU2489
slr0773 (mRNA / CDS) - TU2492
slr0773 (Protein) - TU2492
slr0782 (mRNA / CDS) - TU2517
slr0782 (Protein) - TU2517
slr0784 (mRNA / CDS) - TU2522
slr0784 (Protein) - TU2522
slr0787 (mRNA / CDS) - TU3211
slr0787 (Protein) - TU3211
slr0787-5UTR (5'UTR) - TU3211
slr0801 (mRNA / CDS) - TU3242
slr0801 (Protein) - TU3242
slr0806 (mRNA / CDS) - TU1725
slr0806 (Protein) - TU1725
slr0815 (mRNA / CDS) - TU1749
slr0815 (Protein) - TU1749
slr0816 (Protein) - TU1749
slr0818 (mRNA / CDS) - TU1754
slr0818 (Protein) - TU1754
slr0821 (mRNA / CDS) - TU1762
slr0821 (Protein) - TU1762
slr0821-3UTR (3'UTR) - TU1762
slr0822 (mRNA / CDS) - TU1765
slr0822 (Protein) - TU1765
slr0822-5UTR (5'UTR) - TU1765
slr0825 (mRNA / CDS) - TU2983
slr0825 (Protein) - TU2983
slr0829 (mRNA / CDS) - TU2988
slr0829 (Protein) - TU2988
slr0841 (mRNA / CDS) - TU3006
slr0841 (Protein) - TU3006
slr0842 (mRNA / CDS) - TU3006
slr0842 (Protein) - TU3006
slr0848 (mRNA / CDS) - TU3024
slr0848 (Protein) - TU3024
slr0852 (mRNA / CDS) - TU1377
slr0852 (Protein) - TU1377
slr0856 (transp mRNA / CDS) - TU1384
slr0863 (mRNA / CDS) - TU1396
slr0863 (Protein) - TU1396
slr0864 (mRNA / CDS) - TU1403
slr0864 (Protein) - TU1403
slr0864-as (asRNA)
slr0865 (mRNA / CDS) - TU1404
slr0865 (Protein) - TU1404
slr0867 (Protein)
slr0869 (mRNA / CDS) - TU1409
slr0869 (Protein) - TU1409
slr0869-as2 (asRNA)
slr0870 (mRNA / CDS) - TU1409
slr0870 (Protein) - TU1409
slr0871 (Protein) - TU1409
slr0872 (mRNA / CDS) - TU1409
slr0872 (Protein) - TU1409
slr0876 (mRNA / CDS) - TU1167
slr0876 (Protein) - TU1167
slr0876-3UTR (3'UTR) - TU1167
slr0878 (mRNA / CDS)
slr0878 (Protein)
slr0880 (mRNA / CDS) - TU1175
slr0880 (Protein) - TU1175
slr0881 (mRNA / CDS) - TU1179
slr0881 (Protein) - TU1179
slr0881-5UTR (5'UTR) - TU1179
slr0885 (mRNA / CDS) - TU1191
slr0885 (Protein) - TU1191
slr0887 (mRNA / CDS) - TU1195
slr0887 (Protein) - TU1195
slr0889 (mRNA / CDS) - TU1196
slr0889 (Protein) - TU1196
slr0890 (mRNA / CDS) - TU1197
slr0890 (Protein) - TU1197
slr0895 (mRNA / CDS)
slr0895 (Protein)
slr0907 (mRNA / CDS) - TU2929
slr0907 (Protein) - TU2929
slr0907-as (asRNA)
slr0909 (mRNA / CDS) - TU2929
slr0909 (Protein) - TU2929
slr0912 (mRNA / CDS) - TU2929
slr0912 (Protein) - TU2929
slr0913 (mRNA / CDS) - TU2929
slr0913 (Protein) - TU2929
slr0914 (mRNA / CDS) - TU2929
slr0914 (Protein) - TU2929
slr0915 (mRNA / CDS) - TU2935
slr0915 (Protein) - TU2935
slr0920 (mRNA / CDS) - TU2937
slr0920 (Protein) - TU2937
slr0924 (mRNA / CDS) - TU2944
slr0924 (Protein) - TU2944
slr0924-3UTR (3'UTR) - TU2944
slr0924-5UTR (5'UTR) - TU2944
slr0929 (mRNA / CDS) - TU3403
slr0929 (Protein) - TU3403
slr0929-5UTR (5'UTR) - TU3403
slr0930 (mRNA / CDS) - TU3403
slr0930 (Protein) - TU3403
slr0935 (mRNA / CDS) - TU2072
slr0935 (Protein) - TU2072
slr0935-5UTR (5'UTR) - TU2072
slr0937 (mRNA / CDS) - TU2074
slr0937 (Protein) - TU2074
slr0938 (mRNA / CDS) - TU2075
slr0938 (Protein) - TU2075
slr0941 (Protein) - TU2078
slr0942 (mRNA / CDS) - TU2079
slr0942 (Protein) - TU2079
slr0948 (mRNA / CDS) - TU2084
slr0948 (Protein) - TU2084
slr0953 (mRNA / CDS) - TU2095
slr0953 (Protein) - TU2095
slr0955 (mRNA / CDS) - TU2098
slr0955 (Protein) - TU2098
slr0959 (mRNA / CDS) - TU2105
slr0959 (Protein) - TU2105
slr0960 (mRNA / CDS) - TU2106
slr0960 (Protein) - TU2106
slr0962 (mRNA / CDS) - TU315
slr0962 (Protein) - TU315
slr0967 (mRNA / CDS) - TU325
slr0967 (Protein) - TU325
slr0967-3UTR (3'UTR) - TU325
slr0967-5UTR (5'UTR) - TU325
slr0971 (mRNA / CDS) - TU330
slr0971 (Protein) - TU330
slr0976 (mRNA / CDS) - TU349
slr0976-5UTR (5'UTR) - TU349
slr0977 (mRNA / CDS) - TU349
slr0977 (Protein) - TU349
slr0981 (mRNA / CDS) - TU352
slr0981 (Protein) - TU352
slr0989 (mRNA / CDS) - TU2349
slr0989 (Protein) - TU2349
slr0990 (mRNA / CDS) - TU2350
slr0990 (Protein) - TU2350
slr0992 (mRNA / CDS) - TU2350
slr0992 (Protein) - TU2350
slr1019 (mRNA / CDS) - TU628
slr1019 (Protein) - TU628
slr1019-3UTR (3'UTR) - TU628
slr1024 (Protein) - TU640
slr1028 (mRNA / CDS) - TU647
slr1028 (Protein) - TU647
slr1028-as8 (asRNA)
slr1032 (Protein) - TU656
slr1033 (mRNA / CDS) - TU656
slr1033 (Protein) - TU656
slr1035 (mRNA / CDS) - TU659
slr1035 (Protein) - TU659
slr1037 (Protein) - TU663
slr1039 (mRNA / CDS) - TU667
slr1039 (Protein) - TU667
slr1041 (mRNA / CDS) - TU479
slr1041 (Protein) - TU479
slr1042 (mRNA / CDS) - TU481
slr1042 (Protein) - TU481
ycf63/slr1045 (mRNA / CDS) - TU481
slr1045,slr1344 (Protein)
slr1046 (Protein) - TU483
slr1048 (mRNA / CDS) - TU484
slr1048 (Protein) - TU484
slr1053 (mRNA / CDS)
slr1053 (Protein)
slr1056 (mRNA / CDS) - TU497
slr1056 (Protein) - TU497
slr1063 (mRNA / CDS) - TU355
slr1063 (Protein) - TU355
slr1065 (mRNA / CDS) - TU355
slr1065 (Protein) - TU355
slr1066 (mRNA / CDS) - TU355
slr1066 (Protein) - TU355
slr1070 (Protein) - TU355
slr1071 (mRNA / CDS) - TU355
slr1071 (Protein) - TU355
slr1073 (mRNA / CDS) - TU355
slr1073 (Protein) - TU355
slr1075 (transp mRNA / CDS) - TU362
slr1076 (mRNA / CDS) - TU362
slr1076 (Protein) - TU362
slr1076-as (asRNA)
slr1077 (mRNA / CDS) - TU364
slr1077 (Protein) - TU364
slr1077-as (asRNA)
slr1079 (mRNA / CDS) - TU366
slr1079 (Protein) - TU366
slr1081 (mRNA / CDS) - TU369
slr1081 (Protein) - TU369
slr1083 (mRNA / CDS) - TU369
slr1083 (Protein) - TU369
slr1084 (mRNA / CDS) - TU369
slr1084 (Protein) - TU369
slr1085 (mRNA / CDS) - TU369
slr1085 (Protein) - TU369
slr1087 (mRNA / CDS) - TU369
slr1087 (Protein) - TU369
slr1094 (mRNA / CDS) - TU183
slr1094 (Protein) - TU183
slr1094-5UTR (5'UTR) - TU183
slr1095 (mRNA / CDS) - TU185
slr1095 (Protein) - TU185
slr1097 (mRNA / CDS) - TU186
slr1097 (Protein) - TU186
slr1098 (mRNA / CDS) - TU186
slr1098 (Protein) - TU186
slr1101 (mRNA / CDS) - TU188
slr1101 (Protein) - TU188
slr1102 (mRNA / CDS) - TU188
slr1102 (Protein) - TU188
slr1102-as2 (asRNA)
slr1103 (mRNA / CDS) - TU191
slr1103 (Protein) - TU191
slr1103-5UTR (5'UTR) - TU191
slr1104 (mRNA / CDS) - TU191
slr1104 (Protein) - TU191
slr1107 (mRNA / CDS) - TU200
slr1107 (Protein) - TU200
slr1110 (mRNA / CDS) - TU212
slr1110 (Protein) - TU212
slr1113 (mRNA / CDS) - TU57
slr1113 (Protein) - TU57
slr1114 (mRNA / CDS) - TU57
slr1114 (Protein) - TU57
slr1115 (mRNA / CDS) - TU57
slr1115 (Protein) - TU57
slr1116 (Protein) - TU57
slr1117 (mRNA / CDS) - TU57
slr1117 (Protein) - TU57
slr1122 (mRNA / CDS) - TU77
slr1122 (Protein) - TU77
slr1127 (mRNA / CDS) - TU84
slr1127 (Protein) - TU84
slr1128 (mRNA / CDS) - TU84
slr1128 (Protein) - TU84
slr1140 (mRNA / CDS) - TU793
slr1140 (Protein) - TU793
slr1142 (mRNA / CDS) - TU800
slr1142 (Protein) - TU800
slr1143 (mRNA / CDS) - TU800
slr1143 (Protein) - TU800
slr1149 (mRNA / CDS) - TU814
slr1149 (Protein) - TU814
slr1150 (mRNA / CDS) - TU814
slr1150 (Protein) - TU814
slr1160 (Protein) - TU1994
slr1161 (mRNA / CDS) - TU1999
slr1161 (Protein) - TU1999
slr1162 (mRNA / CDS) - TU1999
slr1162 (Protein) - TU1999
slr1166 (mRNA / CDS) - TU2006
slr1166 (Protein) - TU2006
slr1169 (mRNA / CDS) - TU2009
slr1169 (Protein) - TU2009
slr1173 (mRNA / CDS) - TU2014
slr1173 (Protein) - TU2014
slr1177 (mRNA / CDS) - TU3679
slr1177 (Protein) - TU3679
slr1178 (mRNA / CDS) - TU3680
slr1178 (Protein) - TU3680
slr1183 (Protein)
slr1186 (mRNA / CDS) - TU3685
slr1186 (Protein) - TU3685
slr1189 (mRNA / CDS) - TU3686
slr1189 (Protein) - TU3686
slr1192 (mRNA / CDS) - TU3687
slr1192 (Protein) - TU3687
slr1194 (mRNA / CDS) - TU881
slr1194 (Protein) - TU881
slr1195 (mRNA / CDS) - TU881
slr1195 (Protein) - TU881
slr1196 (mRNA / CDS) - TU881
slr1196 (Protein) - TU881
smf/slr1197 (mRNA / CDS) - TU881
slr1197 (Protein) - TU881
slr1198 (mRNA / CDS) - TU886
slr1198 (Protein) - TU886
slr1198-5UTR (5'UTR) - TU886
slr1201 (mRNA / CDS) - TU888
slr1201 (Protein) - TU888
slr1203 (mRNA / CDS) - TU890
slr1203 (Protein) - TU890
slr1205 (mRNA / CDS) - TU893
slr1205 (Protein) - TU893
slr1205-5UTR (5'UTR) - TU893
slr1206 (mRNA / CDS) - TU893
slr1206 (Protein) - TU893
slr1207 (mRNA / CDS) - TU893
slr1207 (Protein) - TU893
slr1208 (mRNA / CDS) - TU896
slr1208 (Protein) - TU896
slr1209 (Protein) - TU896
slr1210 (Protein) - TU896
slr1220 (mRNA / CDS) - TU1041
slr1220 (Protein) - TU1041
slr1232 (mRNA / CDS) - TU1063
slr1232 (Protein) - TU1063
slr1232-3UTR (3'UTR) - TU1063
slr1232-5UTR (5'UTR) - TU1063
slr1234 (mRNA / CDS) - TU1069
slr1234 (Protein) - TU1069
slr1235 (mRNA / CDS) - TU1070
slr1235 (Protein) - TU1070
slr1236 (mRNA / CDS) - TU1070
slr1236 (Protein) - TU1070
slr1240 (mRNA / CDS) - TU1089
slr1240 (Protein) - TU1089
slr1241 (mRNA / CDS) - TU1089
slr1241 (Protein) - TU1089
slr1253 (mRNA / CDS) - TU1437
slr1253 (Protein) - TU1437
slr1253-3UTR (3'UTR) - TU1437
slr1253-5UTR (5'UTR) - TU1437
slr1257 (mRNA / CDS)
slr1257 (Protein)
slr1258 (mRNA / CDS) - TU1445
slr1258 (Protein) - TU1445
slr1258-5UTR (5'UTR) - TU1445
slr1259 (mRNA / CDS) - TU1446
slr1259 (Protein) - TU1446
slr1260 (mRNA / CDS) - TU1446
slr1260 (Protein) - TU1446
slr1261 (mRNA / CDS) - TU1446
slr1261 (Protein) - TU1446
slr1263 (mRNA / CDS) - TU1448
slr1263 (Protein) - TU1448
slr1263-3UTR (3'UTR) - TU1448
slr1263-as (asRNA)
slr1266 (mRNA / CDS) - TU1938
slr1266 (Protein) - TU1938
slr1270 (mRNA / CDS) - TU1942
slr1270 (Protein) - TU1942
slr1270-5UTR (5'UTR) - TU1942
slr1272 (mRNA / CDS) - TU1945
slr1272 (Protein) - TU1945
slr1272-5UTR (5'UTR) - TU1945
slr1273 (mRNA / CDS) - TU1945
slr1273 (Protein) - TU1945
slr1275 (mRNA / CDS) - TU1945
slr1275 (Protein) - TU1945
slr1276 (mRNA / CDS) - TU1945
slr1276 (Protein) - TU1945
slr1287 (mRNA / CDS) - TU1960
slr1287 (Protein) - TU1960
slr1288 (mRNA / CDS) - TU1960
slr1288 (Protein) - TU1960
slr1290 (mRNA / CDS) - TU275
slr1290 (Protein) - TU275
slr1290-as (asRNA)
slr1299 (mRNA / CDS) - TU295
slr1299 (Protein) - TU295
slr1300 (mRNA / CDS) - TU296
slr1300 (Protein) - TU296
slr1301 (mRNA / CDS) - TU302
slr1301 (Protein) - TU302
slr1305 (mRNA / CDS) - TU306
slr1305 (Protein) - TU306
slr1312 (mRNA / CDS) - TU14
slr1312 (Protein) - TU14
slr1315 (mRNA / CDS) - TU17
slr1315 (Protein) - TU17
slr1334 (mRNA / CDS) - TU1717
slr1334 (Protein) - TU1717
slr1336 (mRNA / CDS) - TU1781
slr1336 (Protein) - TU1781
slr1338 (Protein) - TU1786
slr1339 (Protein) - TU1786
slr1340 (Protein) - TU1786
slr1342 (mRNA / CDS) - TU1790
slr1342 (Protein) - TU1790
slr1343 (mRNA / CDS) - TU1790
slr1343 (Protein) - TU1790
slr1344 (mRNA / CDS) - TU1795
slr1353 (mRNA / CDS) - TU1810
slr1353 (Protein) - TU1810
slr1357 (transp mRNA / CDS) - TU1129
slr1362 (mRNA / CDS) - TU1134
slr1362 (Protein) - TU1134
slr1363 (mRNA / CDS) - TU1136
slr1363 (Protein) - TU1136
slr1378 (mRNA / CDS) - TU672
slr1378 (Protein) - TU672
slr1383 (mRNA / CDS) - TU674
slr1383 (Protein) - TU674
slr1384 (mRNA / CDS) - TU674
slr1384 (Protein) - TU674
slr1385 (mRNA / CDS) - TU677
slr1385 (Protein) - TU677
slr1391 (mRNA / CDS) - TU685
slr1391 (Protein) - TU685
slr1394 (mRNA / CDS) - TU695
slr1394 (Protein) - TU695
slr1394-5UTR (5'UTR) - TU695
slr1395 (Protein) - TU697
slr1397 (mRNA / CDS) - TU700
slr1397 (Protein) - TU700
slr1398 (mRNA / CDS) - TU701
slr1398 (Protein) - TU701
slr1403 (mRNA / CDS) - TU1209
slr1403 (Protein) - TU1209
slr1406 (mRNA / CDS) - TU1223
slr1406 (Protein) - TU1223
slr1407 (mRNA / CDS) - TU1223
slr1407 (Protein) - TU1223
slr1409 (mRNA / CDS) - TU1223
slr1409 (Protein) - TU1223
slr1410 (mRNA / CDS) - TU1223
slr1410 (Protein) - TU1223
slr1411 (mRNA / CDS) - TU165
slr1411 (Protein) - TU165
slr1413 (mRNA / CDS) - TU169
slr1413 (Protein) - TU169
slr1415 (mRNA / CDS) - TU170
slr1415 (Protein) - TU170
slr1419 (mRNA / CDS) - TU3438
slr1419 (Protein) - TU3438
slr1420 (mRNA / CDS) - TU3438
slr1420 (Protein) - TU3438
slr1421 (mRNA / CDS) - TU3439
slr1421 (Protein) - TU3439
slr1425 (mRNA / CDS) - TU3448
slr1425,ssr5092 (Protein)
slr1428 (mRNA / CDS) - TU3460
slr1428 (Protein) - TU3460
slr1431 (mRNA / CDS) - TU3467
slr1431 (Protein) - TU3467
slr1431-3UTR (3'UTR) - TU3467
slr1437 (mRNA / CDS) - TU1091
slr1437 (Protein) - TU1091
slr1437-5UTR (5'UTR) - TU1091
slr1438 (mRNA / CDS) - TU1091
slr1438 (Protein) - TU1091
slr1440 (mRNA / CDS) - TU1098
slr1440 (Protein) - TU1098
slr1450 (mRNA / CDS) - TU1120
slr1450 (Protein) - TU1120
slr1462 (mRNA / CDS) - TU3488
slr1462 (Protein) - TU3488
slr1462-5UTR (5'UTR) - TU3488
slr1464 (mRNA / CDS) - TU3495
slr1464 (Protein) - TU3495
slr1467 (mRNA / CDS) - TU1895
slr1467 (Protein) - TU1895
slr1467-as (asRNA)
slr1468 (Protein)
slr1470 (mRNA / CDS) - TU1898
slr1470 (Protein) - TU1898
slr1471 (mRNA / CDS) - TU1898
slr1471 (Protein) - TU1898
slr1472 (mRNA / CDS) - TU1898
slr1472 (Protein) - TU1898
slr1474 (mRNA / CDS) - TU1905
slr1474 (Protein) - TU1905
slr1478 (mRNA / CDS) - TU1913
slr1478 (Protein) - TU1913
slr1488 (mRNA / CDS) - TU37
slr1488 (Protein) - TU37
slr1501 (mRNA / CDS) - TU55
slr1501 (Protein) - TU55
slr1505 (mRNA / CDS) - TU1628
slr1505 (Protein) - TU1628
slr1506 (mRNA / CDS) - TU1632
slr1506 (Protein) - TU1632
slr1507 (mRNA / CDS) - TU1633
slr1507 (Protein) - TU1633
slr1507-5UTR (5'UTR) - TU1633
slr1513 (mRNA / CDS) - TU1642
slr1513 (Protein) - TU1642
slr1522 (transp mRNA / CDS) - TU1655
slr1524 (transp mRNA / CDS) - TU1659
slr1529 (mRNA / CDS)
slr1529 (Protein)
slr1533 (Protein) - TU1980
slr1534 (mRNA / CDS) - TU1980
slr1534 (Protein) - TU1980
slr1537 (mRNA / CDS) - TU994
slr1537 (Protein) - TU994
slr1540 (mRNA / CDS) - TU995
slr1540 (Protein) - TU995
slr1540-5UTR (5'UTR) - TU995
slr1541 (mRNA / CDS) - TU995
slr1541 (Protein) - TU995
slr1546 (mRNA / CDS) - TU1002
slr1546 (Protein) - TU1002
slr1547 (mRNA / CDS) - TU1002
slr1547 (Protein) - TU1002
slr1552 (mRNA / CDS) - TU1019
slr1552 (Protein) - TU1019
slr1557 (mRNA / CDS) - TU3540
slr1557 (Protein) - TU3540
slr1563 (mRNA / CDS) - TU3549
slr1563 (Protein) - TU3549
slr1565 (mRNA / CDS) - TU3554
slr1565 (Protein) - TU3554
slr1568 (Protein)
slr1570 (mRNA / CDS) - TU3560
slr1570 (Protein) - TU3560
slr1570-5UTR (5'UTR) - TU3560
slr1571 (mRNA / CDS) - TU3560
slr1571 (Protein) - TU3560
slr1572 (mRNA / CDS) - TU3560
slr1572 (Protein) - TU3560
slr1573 (mRNA / CDS) - TU3562
slr1573 (Protein) - TU3562
slr1573-3UTR (3'UTR) - TU3562
slr1575 (mRNA / CDS) - TU3565
slr1575 (Protein) - TU3565
slr1576 (mRNA / CDS) - TU3567
slr1576 (Protein) - TU3567
slr1577 (mRNA / CDS) - TU3567
slr1577 (Protein) - TU3567
slr1579 (mRNA / CDS) - TU3568
slr1579 (Protein) - TU3568
slr1579-3UTR (3'UTR) - TU3568
slr1584 (mRNA / CDS) - TU3582
slr1584 (Protein) - TU3582
slr1585 (transp mRNA / CDS) - TU3582
slr1588 (mRNA / CDS) - TU3586
slr1588 (Protein) - TU3586
slr1590 (mRNA / CDS) - TU430
slr1590 (Protein) - TU430
slr1597 (mRNA / CDS) - TU443
slr1597 (Protein) - TU443
slr1599 (mRNA / CDS) - TU451
slr1599 (Protein) - TU451
slr1599-5UTR (5'UTR) - TU451
slr1600 (mRNA / CDS) - TU454
slr1600 (Protein) - TU454
slr1600-3UTR (3'UTR) - TU454
slr1603 (mRNA / CDS) - TU459
slr1603 (Protein) - TU459
slr1609 (mRNA / CDS) - TU476
slr1609 (Protein) - TU476
slr1610 (mRNA / CDS) - TU353
slr1610 (Protein) - TU353
slr1611 (mRNA / CDS) - TU353
slr1611 (Protein) - TU353
slr1612 (mRNA / CDS) - TU355
slr1612,slr0976 (Protein)
slr1612-as (asRNA)
slr1613 (mRNA / CDS) - TU355
slr1613 (Protein) - TU355
slr1614 (mRNA / CDS) - TU355
slr1614 (Protein) - TU355
slr1616 (mRNA / CDS) - TU355
slr1616 (Protein) - TU355
slr1617 (mRNA / CDS) - TU355
slr1617 (Protein) - TU355
slr1618 (mRNA / CDS) - TU355
slr1618 (Protein) - TU355
slr1619 (mRNA / CDS) - TU355
slr1619 (Protein) - TU355
slr1624 (mRNA / CDS) - TU2113
slr1624 (Protein) - TU2113
slr1624-as (asRNA)
slr1626 (mRNA / CDS) - TU2118
slr1626 (Protein) - TU2118
slr1628 (mRNA / CDS) - TU2122
slr1628 (Protein) - TU2122
slr1629 (mRNA / CDS) - TU2122
slr1629 (Protein) - TU2122
slr1634 (mRNA / CDS) - TU2127
slr1634 (Protein) - TU2127
slr1634-3UTR (3'UTR) - TU2127
slr1634-5UTR (5'UTR) - TU2127
slr1635 (transp mRNA / CDS) - TU2129
slr1636 (mRNA / CDS) - TU2129
slr1636 (Protein) - TU2129
slr1644 (mRNA / CDS) - TU2147
slr1644 (Protein) - TU2147
slr1647 (mRNA / CDS) - TU2148
slr1647 (Protein) - TU2148
slr1648 (mRNA / CDS) - TU2150
slr1648 (Protein) - TU2150
slr1657 (mRNA / CDS) - TU3638
slr1657 (Protein) - TU3638
slr1658 (Protein) - TU3638
slr1659 (Protein) - TU3638
slr1660 (mRNA / CDS) - TU3640
slr1660 (Protein) - TU3640
slr1661 (mRNA / CDS) - TU3645
slr1661 (Protein) - TU3645
slr1664 (mRNA / CDS) - TU3648
slr1664 (Protein) - TU3648
slr1667 (mRNA / CDS) - TU3658
slr1667 (Protein) - TU3658
slr1667-3UTR (3'UTR) - TU3658
slr1667-5UTR (5'UTR) - TU3658
slr1668 (mRNA / CDS) - TU3661
slr1668 (Protein) - TU3661
slr1668-5UTR (5'UTR) - TU3661
slr1670 (mRNA / CDS)
slr1670 (Protein)
slr1677 (mRNA / CDS) - TU2026
slr1677 (Protein) - TU2026
slr1681 (mRNA / CDS) - TU2036
slr1681 (Protein) - TU2036
slr1682 (transp mRNA / CDS) - TU2036
slr1683 (transp mRNA / CDS) - TU2041
slr1686 (mRNA / CDS) - TU2045
slr1686 (Protein) - TU2045
slr1686-3UTR (3'UTR) - TU2045
slr1687 (mRNA / CDS) - TU2046
slr1687 (Protein) - TU2046
slr1687-3UTR (3'UTR) - TU2046
slr1690 (Protein) - TU2057
slr1692 (mRNA / CDS) - TU2062
slr1692 (Protein) - TU2062
slr1693 (mRNA / CDS) - TU2063
slr1693 (Protein) - TU2063
slr1694 (mRNA / CDS) - TU2063
slr1694 (Protein) - TU2063
slr1699 (mRNA / CDS)
slr1699 (Protein)
slr1702 (mRNA / CDS) - TU712
slr1702 (Protein) - TU712
slr1704 (mRNA / CDS) - TU725
slr1704 (Protein) - TU725
slr1704-3UTR (3'UTR) - TU725
slr1704-5UTR (5'UTR) - TU725
slr1704-as (asRNA)
slr1712 (mRNA / CDS) - TU1490
slr1712 (Protein) - TU1490
slr1715 (transp mRNA / CDS) - TU1498
slr1717 (mRNA / CDS) - TU1502
slr1717 (Protein) - TU1502
slr1721 (mRNA / CDS) - TU1506
slr1721 (Protein) - TU1506
slr1724 (mRNA / CDS) - TU1513
slr1724 (Protein) - TU1513
slr1732 (mRNA / CDS) - TU1345
slr1732 (Protein) - TU1345
slr1732-3UTR (3'UTR) - TU1345
slr1740 (mRNA / CDS) - TU1363
slr1740 (Protein) - TU1363
slr1740-5UTR (5'UTR) - TU1363
slr1742 (mRNA / CDS) - TU1366
slr1742 (Protein) - TU1366
slr1748 (mRNA / CDS) - TU1029
slr1748 (Protein) - TU1029
slr1752 (mRNA / CDS) - TU1035
slr1752 (Protein) - TU1035
slr1753 (mRNA / CDS) - TU505
slr1753 (Protein) - TU505
slr1753-3UTR (3'UTR) - TU505
slr1760 (mRNA / CDS) - TU513
slr1760 (Protein) - TU513
slr1762 (mRNA / CDS) - TU516
slr1762 (Protein) - TU516
slr1762-5UTR (5'UTR) - TU516
slr1763 (mRNA / CDS) - TU517
slr1763 (Protein) - TU517
slr1767 (Protein)
slr1768 (mRNA / CDS) - TU525
slr1768 (Protein) - TU525
slr1771 (mRNA / CDS) - TU528
slr1771 (Protein) - TU528
slr1772 (mRNA / CDS) - TU212
slr1772 (Protein) - TU212
slr1773 (Protein)
slr1776 (mRNA / CDS) - TU216
slr1776 (Protein) - TU216
slr1778 (Protein) - TU221
slr1788 (mRNA / CDS) - TU235
slr1788 (Protein) - TU235
slr1789 (mRNA / CDS) - TU235
slr1789 (Protein) - TU235
slr1790 (mRNA / CDS) - TU238
slr1790 (Protein) - TU238
slr1794 (mRNA / CDS) - TU246
slr1794 (Protein) - TU246
slr1796 (mRNA / CDS) - TU249
slr1796 (Protein) - TU249
slr1796-3UTR (3'UTR) - TU249
slr1799 (mRNA / CDS) - TU264
slr1799 (Protein) - TU264
slr1800 (mRNA / CDS) - TU266
slr1800 (Protein) - TU266
slr1809 (mRNA / CDS) - TU1321
slr1809 (Protein) - TU1321
slr1809-5UTR (5'UTR) - TU1321
slr1811 (mRNA / CDS) - TU1325
slr1811 (Protein) - TU1325
slr1812 (mRNA / CDS) - TU1325
slr1812 (Protein) - TU1325
slr1813 (mRNA / CDS) - TU1326
slr1813 (Protein) - TU1326
slr1814 (mRNA / CDS) - TU1326
slr1814 (Protein) - TU1326
slr1816 (mRNA / CDS) - TU1328
slr1816 (Protein) - TU1328
slr1818 (mRNA / CDS) - TU1334
slr1818 (Protein) - TU1334
slr1818-3UTR (3'UTR) - TU1334
slr1819 (mRNA / CDS) - TU1336
slr1819 (Protein) - TU1336
slr1820 (mRNA / CDS)
slr1820 (Protein)
slr1821 (mRNA / CDS) - TU1344
slr1821 (Protein) - TU1344
slr1826 (Protein) - TU936
slr1837 (mRNA / CDS) - TU965
slr1837 (Protein) - TU965
slr1841 (mRNA / CDS) - TU971
slr1841 (Protein) - TU971
slr1841-3UTR (3'UTR) - TU971
slr1841-5UTR (5'UTR) - TU971
slr1847 (mRNA / CDS) - TU534
slr1847 (Protein) - TU534
slr1847-as (asRNA)
slr1852 (mRNA / CDS) - TU1227
slr1852 (Protein) - TU1227
slr1853 (mRNA / CDS) - TU1227
slr1853 (Protein) - TU1227
slr1854 (Protein) - TU1227
slr1855 (mRNA / CDS) - TU1227
slr1855 (Protein) - TU1227
slr1856 (mRNA / CDS) - TU1227
slr1856 (Protein) - TU1227
slr1859 (mRNA / CDS) - TU1227
slr1859 (Protein) - TU1227
slr1861 (mRNA / CDS) - TU1227
slr1861 (Protein) - TU1227
slr1862 (Protein) - TU1230
slr1863 (mRNA / CDS) - TU1232
slr1863 (Protein) - TU1232
slr1865 (mRNA / CDS) - TU1233
slr1865 (Protein) - TU1233
slr1866 (mRNA / CDS) - TU1233
slr1866 (Protein) - TU1233
slr1869 (mRNA / CDS) - TU1242
slr1869 (Protein) - TU1242
slr1870 (Protein) - TU1245
slr1876 (Protein)
slr1880 (Protein)
slr1888 (mRNA / CDS) - TU1282
slr1888 (Protein) - TU1282
slr1894 (mRNA / CDS) - TU848
slr1894 (Protein) - TU848
slr1894-3UTR (3'UTR) - TU848
slr1894-5UTR (5'UTR) - TU848
slr1895 (mRNA / CDS) - TU850
slr1895 (Protein) - TU850
slr1895-5UTR (5'UTR) - TU850
slr1896 (mRNA / CDS) - TU852
slr1896 (Protein) - TU852
slr1900 (mRNA / CDS) - TU858
slr1900 (Protein) - TU858
slr1901 (mRNA / CDS) - TU858
slr1901 (Protein) - TU858
slr1902 (transp mRNA / CDS) - TU860
slr1907 (mRNA / CDS) - TU598
slr1907 (Protein) - TU598
slr1908 (mRNA / CDS) - TU599
slr1908 (Protein) - TU599
slr1908-5UTR (5'UTR) - TU599
slr1909 (mRNA / CDS) - TU599
slr1909 (Protein) - TU599
slr1911 (Protein) - TU603
slr1911-as (asRNA)
slr1912 (mRNA / CDS) - TU604
slr1912 (Protein) - TU604
slr1915 (mRNA / CDS) - TU611
slr1915 (Protein) - TU611
slr1916 (mRNA / CDS) - TU612
slr1916 (Protein) - TU612
slr1916-5UTR (5'UTR) - TU612
slr1917 (mRNA / CDS) - TU613
slr1917 (Protein) - TU613
slr1917-3UTR (3'UTR) - TU613
slr1918 (mRNA / CDS) - TU615
slr1918 (Protein) - TU615
slr1918-3UTR (3'UTR) - TU615
slr1919 (mRNA / CDS) - TU622
slr1919 (Protein) - TU622
slr1920 (mRNA / CDS) - TU624
slr1920 (Protein) - TU624
slr1920-3UTR (3'UTR) - TU624
slr1923 (mRNA / CDS) - TU2293
slr1923 (Protein) - TU2293
slr1924 (mRNA / CDS) - TU2293
slr1924 (Protein) - TU2293
slr1926 (mRNA / CDS) - TU2299
slr1926 (Protein) - TU2299
slr1927 (mRNA / CDS) - TU2299
slr1927 (Protein) - TU2299
slr1932 (mRNA / CDS) - TU2301
slr1932 (Protein) - TU2301
slr1936 (transp mRNA / CDS) - TU2323
slr1937 (transp mRNA / CDS) - TU2323
slr1939 (mRNA / CDS) - TU2325
slr1939 (Protein) - TU2325
slr1940 (mRNA / CDS) - TU2331
slr1940 (Protein) - TU2331
slr1944 (mRNA / CDS) - TU2336
slr1944 (Protein) - TU2336
slr1946 (mRNA / CDS) - TU2342
slr1946 (Protein) - TU2342
slr1949 (mRNA / CDS) - TU2346
slr1949 (Protein) - TU2346
slr1951 (mRNA / CDS) - TU2348
slr1951 (Protein) - TU2348
slr1958 (mRNA / CDS) - TU1816
slr1958 (Protein) - TU1816
slr1959 (mRNA / CDS) - TU1819
slr1959 (Protein) - TU1819
slr1960 (transp mRNA / CDS) - TU1822
slr1962 (mRNA / CDS) - TU1824
slr1962 (Protein) - TU1824
slr1963 (mRNA / CDS) - TU1827
slr1963 (Protein) - TU1827
slr1963-5UTR (5'UTR) - TU1827
slr1964 (mRNA / CDS) - TU1827
slr1964 (Protein) - TU1827
slr1968 (mRNA / CDS) - TU1833
slr1968 (Protein) - TU1833
slr1970 (mRNA / CDS) - TU1845
slr1970 (Protein) - TU1845
slr1971 (mRNA / CDS) - TU1847
slr1971 (Protein) - TU1847
slr1975 (mRNA / CDS) - TU1864
slr1975 (Protein) - TU1864
slr1977 (mRNA / CDS) - TU1868
slr1977 (Protein) - TU1868
slr1978 (mRNA / CDS) - TU1869
slr1978 (Protein) - TU1869
slr1982 (mRNA / CDS) - TU1875
slr1982 (Protein) - TU1875
slr1998 (mRNA / CDS)
slr1998 (Protein)
slr2000 (Protein) - TU1482
slr2004 (mRNA / CDS) - TU1490
slr2004 (Protein) - TU1490
slr2005 (mRNA / CDS) - TU754
slr2005 (Protein) - TU754
slr2006 (mRNA / CDS) - TU756
slr2006 (Protein) - TU756
slr2011 (mRNA / CDS) - TU757
slr2011 (Protein) - TU757
slr2013 (mRNA / CDS) - TU757
slr2013 (Protein) - TU757
slr2018 (mRNA / CDS) - TU763
slr2018 (Protein) - TU763
slr2019 (mRNA / CDS) - TU763
slr2019 (Protein) - TU763
slr2024 (mRNA / CDS) - TU770
slr2024 (Protein) - TU770
slr2025 (mRNA / CDS) - TU771
slr2025 (Protein) - TU771
slr2025-3UTR (3'UTR) - TU771
slr2030 (mRNA / CDS) - TU786
slr2030 (Protein) - TU786
slr2036 (transp mRNA / CDS) - TU554
slr2046 (mRNA / CDS) - TU577
slr2046 (Protein) - TU577
slr2048 (mRNA / CDS) - TU582
slr2048 (Protein) - TU582
slr2052 (mRNA / CDS) - TU585
slr2052 (Protein) - TU585
slr2052-3UTR (3'UTR) - TU585
slr2052-5UTR (5'UTR) - TU585
slr2053 (mRNA / CDS) - TU587
slr2053 (Protein) - TU587
slr2059 (mRNA / CDS) - TU1455
slr2059 (Protein) - TU1455
slr2060 (mRNA / CDS) - TU1455
slr2060 (Protein) - TU1455
slr2062 (transp mRNA / CDS) - TU1460
slr2070 (mRNA / CDS) - TU911
slr2070 (Protein) - TU911
slr2071 (mRNA / CDS) - TU911
slr2071 (Protein) - TU911
slr2077 (mRNA / CDS) - TU926
slr2077 (Protein) - TU926
slr2080 (Protein) - TU1571
slr2084 (mRNA / CDS) - TU1573
slr2084 (Protein) - TU1573
slr2095 (transp mRNA / CDS) - TU1600
slr2100 (mRNA / CDS) - TU1605
slr2100 (Protein) - TU1605
slr2101 (mRNA / CDS) - TU1606
slr2101 (Protein) - TU1606
slr2105 (mRNA / CDS) - TU1611
slr2105 (Protein) - TU1611
slr2112 (transp mRNA / CDS) - TU1672
slr2119 (mRNA / CDS) - TU1674
slr2119 (Protein) - TU1674
slr2120 (mRNA / CDS) - TU1675
slr2120 (Protein) - TU1675
slr2122 (Protein) - TU1677
slr2123 (mRNA / CDS) - TU1677
slr2123 (Protein) - TU1677
slr2124 (mRNA / CDS) - TU1677
slr2124 (Protein) - TU1677
slr2125 (mRNA / CDS) - TU1677
slr2125 (Protein) - TU1677
slr2127 (mRNA / CDS)
slr2127 (Protein)
slr2128 (mRNA / CDS) - TU1679
slr2128 (Protein) - TU1679
slr2131 (mRNA / CDS) - TU1683
slr2131 (Protein) - TU1683
slr2141 (mRNA / CDS)
slr2141 (Protein)
slr2144 (mRNA / CDS) - TU1308
slr2144 (Protein) - TU1308
slr5005 (mRNA / CDS) - TU5005
slr5005 (Protein) - TU5005
slr5005-5UTR (5'UTR) - TU5005
slr5017 (Protein) - TU5024
slr5018 (Protein) - TU5024
slr5022 (mRNA / CDS) - TU5025
slr5022 (Protein) - TU5025
slr5023 (mRNA / CDS)
slr5023 (Protein)
slr5024 (Protein)
slr5029 (transp mRNA / CDS) - TU5034
slr5040 (transp mRNA / CDS) - TU5042
slr5051 (mRNA / CDS) - TU5057
slr5051 (Protein) - TU5057
slr5051-3UTR (3'UTR) - TU5057
slr5054 (mRNA / CDS) - TU5060
slr5054 (Protein) - TU5060
slr5058 (mRNA / CDS) - TU5067
slr5058 (Protein) - TU5067
slr5071 (mRNA / CDS) - TU5086
slr5071 (Protein) - TU5086
slr5071-3UTR (3'UTR) - TU5086
slr5073 (mRNA / CDS) - TU5087
slr5073 (Protein) - TU5087
slr5077 (mRNA / CDS)
slr5077 (Protein)
slr5078 (mRNA / CDS) - TU5091
slr5078 (Protein) - TU5091
slr5078-5UTR (5'UTR) - TU5091
slr5082 (mRNA / CDS) - TU5094
slr5082 (Protein) - TU5094
slr5088 (Protein)
slr5105,sll6036 (Protein)
slr5110 (mRNA / CDS) - TU5115
slr5110 (Protein) - TU5115
slr5110-3UTR (3'UTR) - TU5115
slr5110-5UTR (5'UTR) - TU5115
slr5111 (Protein) - TU5117
slr5118 (mRNA / CDS) - TU5121
slr5118 (Protein) - TU5121
slr5119 (mRNA / CDS) - TU5122
slr5119 (Protein) - TU5122
slr5119-5UTR (5'UTR) - TU5122
slr5124 (mRNA / CDS)
slr5124 (Protein)
slr5126 (Protein) - TU5126
slr5126-5UTR (5'UTR) - TU5126
slr5127 (Protein) - TU5127
slr6012 (mRNA / CDS) - TU6009
slr6014 (mRNA / CDS) - TU6009
slr6015 (mRNA / CDS) - TU6009
slr6037,sll5104 (Protein)
slr6039 (mRNA / CDS) - TU6037
slr6039-5UTR (5'UTR) - TU6037
slr6040 (mRNA / CDS) - TU6037
slr6041 (mRNA / CDS) - TU6037
slr6041,sll0790 (Protein)
slr6042 (mRNA / CDS) - TU6040
slr6042 (Protein) - TU6040
slr6044 (Protein) - TU6042
slr6047 (mRNA / CDS) - TU6044
slr6047 (Protein) - TU6044
slr6049 (mRNA / CDS) - TU6047
slr6049 (Protein) - TU6047
slr6050 (mRNA / CDS) - TU6047
slr6050 (Protein) - TU6047
slr6051 (Protein) - TU6047
slr6056 (Protein) - TU6050
slr6071 (mRNA / CDS) - TU6064
slr6071,slr6012 (Protein)
slr6073 (mRNA / CDS) - TU6064
slr6073,slr6014 (Protein)
slr6074 (mRNA / CDS) - TU6064
slr6074,slr6015 (Protein)
slr6095 (mRNA / CDS) - TU6088
slr6095 (Protein) - TU6088
slr6096 (mRNA / CDS) - TU6088
slr6096 (Protein) - TU6088
slr6097 (mRNA / CDS) - TU6090
slr6097 (Protein) - TU6090
slr6097-5UTR (5'UTR) - TU6090
slr6100 (mRNA / CDS) - TU6093
slr6100 (Protein) - TU6093
slr6101 (Protein) - TU6093
slr6102 (mRNA / CDS) - TU6093
slr6102 (Protein) - TU6093
slr6110 (mRNA / CDS) - TU6104
slr6110 (Protein) - TU6104
slr6110-3UTR (3'UTR) - TU6104
slr7008 (transp mRNA / CDS)
slr7011 (mRNA / CDS) - TU7004
slr7011 (Protein) - TU7004
slr7012 (mRNA / CDS) - TU7004
slr7012 (Protein) - TU7004
slr7013 (mRNA / CDS)
slr7013 (Protein)
slr7024 (mRNA / CDS) - TU7011
slr7024 (Protein) - TU7011
slr7041 (mRNA / CDS) - TU7034
slr7041 (Protein) - TU7034
slr7060 (mRNA / CDS)
slr7060 (Protein)
slr7061 (mRNA / CDS) - TU7055
slr7061 (Protein) - TU7055
slr7061-5UTR (5'UTR) - TU7055
slr7073 (mRNA / CDS) - TU7069
slr7073 (Protein) - TU7069
slr7073-3UTR (3'UTR) - TU7069
slr7080 (mRNA / CDS) - TU7075
slr7080 (Protein) - TU7075
slr7080-5UTR (5'UTR) - TU7075
slr7094 (mRNA / CDS) - TU7087
slr7094 (Protein) - TU7087
slr7094-5UTR (5'UTR) - TU7087
slr7096 (mRNA / CDS) - TU7088
slr7096 (Protein) - TU7088
slr7099 (mRNA / CDS)
slr7099 (Protein)
slr7104 (transp mRNA / CDS) - TU7097
transposases (Protein)
slr7105 (transp mRNA / CDS) - TU7097
slr8015 (mRNA / CDS) - TU8009
slr8015 (Protein) - TU8009
slr8022 (Protein)
slr8023 (mRNA / CDS)
slr8023 (Protein)
slr8030 (mRNA / CDS) - TU8026
slr8030 (Protein) - TU8026
slr8045 (transp mRNA / CDS) - TU8044
slr9101 (Protein)
ssl0105 (Protein)
ssl0241 (Protein) - TU2289
ssl0242 (Protein) - TU2288
ssl0258 (Protein)
ssl0259 (Protein) - TU2279
ssl0294 (mRNA / CDS) - TU2421
ssl0294 (Protein) - TU2421
ssl0294-3UTR (3'UTR) - TU2421
ssl0312 (Protein) - TU2397
ssl0350 (Protein) - TU2896
ssl0352 (mRNA / CDS) - TU2895
ssl0352 (Protein) - TU2895
ssl0385 (Protein) - TU2637
ssl0410 (mRNA / CDS) - TU145
ssl0410 (Protein) - TU145
ssl0410-3UTR (3'UTR) - TU145
ssl0438 (Protein) - TU1569
ssl0461 (mRNA / CDS) - TU1554
ssl0461 (Protein) - TU1554
ssl0467 (Protein) - TU1542
ssl0467-3UTR (3'UTR) - TU1542
ssl0483 (mRNA / CDS) - TU2208
ssl0483 (Protein) - TU2208
ssl0483-5UTR (5'UTR) - TU2208
ssl0511 (mRNA / CDS) - TU3206
ssl0511 (Protein) - TU3206
ssl0511-3UTR (3'UTR) - TU3206
ssl0564 (Protein) - TU2373
ssl0787 (mRNA / CDS) - TU2669
ssl0787 (Protein) - TU2669
ssl0788 (mRNA / CDS) - TU2669
ssl0788 (Protein) - TU2669
ssl0832 (mRNA / CDS)
ssl0832 (Protein)
ssl1046 (mRNA / CDS) - TU2962
ssl1046 (Protein) - TU2962
ssl1255 (Protein) - TU3291
ssl1263 (mRNA / CDS) - TU3286
ssl1263 (Protein) - TU3286
ssl1263-5UTR (5'UTR) - TU3286
ssl1328 (Protein)
ssl1377 (Protein) - TU3603
ssl1378 (mRNA / CDS)
ssl1378 (Protein)
ssl1464 (Protein) - TU3238
ssl1498 (mRNA / CDS) - TU1775
ssl1498 (Protein) - TU1775
ssl1533 (mRNA / CDS) - TU3014
ssl1533 (Protein) - TU3014
ssl1533-3UTR (3'UTR) - TU3014
ssl1533-5UTR (5'UTR) - TU3014
ssl1552 (mRNA / CDS) - TU2996
ssl1552 (Protein) - TU2996
ssl1707 (mRNA / CDS) - TU3409
ssl1707 (Protein) - TU3409
ssl1762 (mRNA / CDS) - TU2070
ssl1762 (Protein) - TU2070
ssl1762-3UTR (3'UTR) - TU2070
ssl1762-5UTR (5'UTR) - TU2070
ssl1792 (Protein) - TU327
ssl1918 (mRNA / CDS) - TU504
ssl1918 (Protein) - TU504
ssl1972 (mRNA / CDS) - TU371
ssl1972 (Protein) - TU371
ssl2009 (mRNA / CDS) - TU198
ssl2009 (Protein) - TU198
ssl2064 (Protein) - TU74
ssl2069 (mRNA / CDS) - TU69
ssl2069 (Protein) - TU69
ssl2069-5UTR (5'UTR) - TU69
ssl2138 (Protein)
ssl2148 (mRNA / CDS) - TU2001
ssl2148 (Protein) - TU2001
ssl2148-3UTR (3'UTR) - TU2001
ssl2245 (mRNA / CDS) - TU1078
ssl2245 (Protein) - TU1078
ssl2420 (mRNA / CDS) - TU1714
ssl2420 (Protein) - TU1714
ssl2501 (mRNA / CDS) - TU1144
ssl2501 (Protein) - TU1144
ssl2501-as (asRNA)
ssl2595 (Protein) - TU1205
ssl2648 (Protein) - TU1106
ssl2717 (mRNA / CDS) - TU1888
ssl2717 (Protein) - TU1888
ssl2733 (mRNA / CDS) - TU46
ssl2733 (Protein) - TU46
ssl2781 (Protein) - TU1631
ssl2807 (mRNA / CDS) - TU1964
ssl2807 (Protein) - TU1964
ssl2814 (mRNA / CDS) - TU1023
ssl2814 (Protein) - TU1023
ssl2823 (Protein) - TU1006
ssl2874 (Protein) - TU3563
ssl2874-3UTR (3'UTR) - TU3563
ssl2920 (mRNA / CDS)
ssl2920 (Protein)
ssl2971 (mRNA / CDS) - TU2139
ssl2971 (Protein) - TU2139
ssl2999 (mRNA / CDS) - TU3657
ssl2999 (Protein) - TU3657
ssl3044 (mRNA / CDS) - TU2044
ssl3044 (Protein) - TU2044
ssl3044-3UTR (3'UTR) - TU2044
ssl3291 (mRNA / CDS) - TU1310
ssl3291 (Protein) - TU1310
ssl3342 (Protein)
ssl3389 (mRNA / CDS) - TU1246
ssl3389 (Protein) - TU1246
ssl3451 (mRNA / CDS) - TU827
ssl3451 (Protein) - TU827
ssl3549 (mRNA / CDS) - TU1849
ssl3549 (Protein) - TU1849
ssl3829 (Protein) - TU1298
ssl5039 (Protein) - TU5041
ssl5095 (Protein) - TU5101
ssl5113 (mRNA / CDS) - TU5119
ssl5113 (Protein) - TU5119
ssl6082,ssl6023 (Protein)
ssl6082-3UTR (3'UTR) - TU6079
ssl7004 (mRNA / CDS) - TU7001
ssl7004 (Protein) - TU7001
ssl7004-5UTR (5'UTR) - TU7001
ssl7038 (Protein) - TU7033
ssl7039 (mRNA / CDS) - TU7033
ssl7039 (Protein) - TU7033
ssl8003 (mRNA / CDS) - TU8005
ssl8003 (Protein) - TU8005
ssl8008 (mRNA / CDS)
ssl8008 (Protein)
ssl8028,ssl7048 (Protein)
ssr0109 (mRNA / CDS) - TU2711
ssr0109 (Protein) - TU2711
ssr0109-5UTR (5'UTR) - TU2711
ssr0332 (mRNA / CDS) - TU2910
ssr0332 (Protein) - TU2910
ssr0332-3UTR (3'UTR) - TU2910
ssr0335 (mRNA / CDS) - TU2913
ssr0335 (Protein) - TU2913
ssr0657 (mRNA / CDS) - TU2235
ssr0657 (Protein) - TU2235
ssr0657-3UTR (3'UTR) - TU2235
ssr0692 (mRNA / CDS) - TU2842
ssr0692 (Protein) - TU2842
ssr0692-3UTR (3'UTR) - TU2842
ssr0692-5UTR (5'UTR) - TU2842
ssr0693 (mRNA / CDS) - TU2844
ssr0693 (Protein) - TU2844
ssr0693-3UTR (3'UTR) - TU2844
ssr0817 (transp mRNA / CDS)
ssr0854 (Protein) - TU3062
ssr1041 (Protein) - TU3119
ssr1049 (mRNA / CDS) - TU2795
ssr1049 (Protein) - TU2795
ssr1049-5UTR (5'UTR) - TU2795
ssr1114 (Protein) - TU3529
ssr1175 (transp mRNA / CDS) - TU3265
ssr1238 (mRNA / CDS)
ssr1238 (Protein)
ssr1256 (mRNA / CDS) - TU3606
ssr1256 (Protein) - TU3606
ssr1260 (Protein) - TU3608
ssr1391 (Protein)
ycf34/ssr1425 (mRNA / CDS) - TU3011
ssr1425 (Protein) - TU3011
ssr1425-3UTR (3'UTR) - TU3011
ssr1473 (Protein) - TU1409
ssr1528 (mRNA / CDS) - TU2926
ssr1528 (Protein) - TU2926
ssr1528-3UTR (3'UTR) - TU2926
ssr1552 (mRNA / CDS) - TU3400
ssr1552 (Protein) - TU3400
ssr1600 (mRNA / CDS) - TU2098
ssr1600 (Protein) - TU2098
ssr1698 (Protein)
ssr1765 (mRNA / CDS) - TU365
ssr1765 (Protein) - TU365
ssr1765-5UTR (5'UTR) - TU365
ssr1766 (mRNA / CDS) - TU365
ssr1766 (Protein) - TU365
ssr1768 (Protein) - TU367
ssr1853 (mRNA / CDS) - TU70
ssr1853 (Protein) - TU70
ssr1951 (mRNA / CDS) - TU2014
ssr1951 (Protein) - TU2014
ssr2047 (mRNA / CDS) - TU1066
ssr2047 (Protein) - TU1066
ssr2062 (mRNA / CDS) - TU1084
ssr2062 (Protein) - TU1084
ssr2062-3UTR (3'UTR) - TU1084
ssr2062-5UTR (5'UTR) - TU1084
ssr2066 (Protein) - TU1089
ssr2067 (mRNA / CDS) - TU1089
ssr2067 (Protein) - TU1089
ssr2201 (Protein) - TU1702
ssr2227 (transp mRNA / CDS) - TU1715
ssr2317 (mRNA / CDS) - TU674
ssr2317 (Protein) - TU674
ssr2406 (mRNA / CDS) - TU1094
ssr2406 (Protein) - TU1094
ssr2422 (mRNA / CDS) - TU1110
ssr2422 (Protein) - TU1110
ssr2422-3UTR (3'UTR) - TU1110
ssr2551 (Protein) - TU1645
ssr2554 (mRNA / CDS) - TU1651
ssr2554 (Protein) - TU1651
ssr2554-5UTR (5'UTR) - TU1651
ssr2754 (Protein) - TU2153
ssr2755 (Protein) - TU2153
ssr2781 (Protein) - TU3651
ssr2787 (mRNA / CDS) - TU3665
ssr2787 (Protein) - TU3665
ssr2962 (Protein) - TU522
ssr2998 (mRNA / CDS) - TU246
ssr2998 (Protein) - TU246
ssr3000 (Protein) - TU246
ssr3122 (mRNA / CDS) - TU532
ssr3122 (Protein) - TU532
ssr3188 (Protein) - TU1272
ssr3189 (mRNA / CDS) - TU1276
ssr3189 (Protein) - TU1276
ssr3189-5UTR (5'UTR) - TU1276
ssr3304 (mRNA / CDS) - TU2336
ssr3304 (Protein) - TU2336
ssr3402 (mRNA / CDS) - TU1490
ssr3402 (Protein) - TU1490
ssr3532 (mRNA / CDS) - TU1571
ssr3532 (Protein) - TU1571
ssr5009 (mRNA / CDS)
ssr5009 (Protein)
ssr6048 (mRNA / CDS) - TU6047
ssr6048 (Protein) - TU6047
TU1005 (NA) - Transcr Unit
TU1007 (NA) - Transcr Unit
TU1009 (NA) - Transcr Unit
TU1027 (NA) - Transcr Unit
TU1032 (NA) - Transcr Unit
TU1034 (NA) - Transcr Unit
TU1038 (NA) - Transcr Unit
TU1042 (NA) - Transcr Unit
TU1045 (NA) - Transcr Unit
TU1046 (NA) - Transcr Unit
TU1050 (NA) - Transcr Unit
TU1056 (NA) - Transcr Unit
TU1080 (NA) - Transcr Unit
TU1086 (NA) - Transcr Unit
TU1087 (NA) - Transcr Unit
TU1088 (NA) - Transcr Unit
TU1093 (NA) - Transcr Unit
TU1119 (NA) - Transcr Unit
TU1121 (NA) - Transcr Unit
TU1145 (NA) - Transcr Unit
TU1148 (NA) - Transcr Unit
TU115 (NA) - Transcr Unit
TU1173 (NA) - Transcr Unit
TU1183 (NA) - Transcr Unit
TU1194 (NA) - Transcr Unit
TU1201 (NA) - Transcr Unit
TU1202 (NA) - Transcr Unit
TU1207 (NA) - Transcr Unit
TU121 (NA) - Transcr Unit
TU1244 (NA) - Transcr Unit
TU1247 (NA) - Transcr Unit
TU125 (NA) - Transcr Unit
TU1257 (NA) - Transcr Unit
TU1268 (NA) - Transcr Unit
TU1274 (NA) - Transcr Unit
TU1284 (NA) - Transcr Unit
TU1285 (NA) - Transcr Unit
TU1294 (NA) - Transcr Unit
TU1299 (NA) - Transcr Unit
TU1302 (NA) - Transcr Unit
TU1311 (NA) - Transcr Unit
TU1313 (NA) - Transcr Unit
TU1315 (NA) - Transcr Unit
TU132 (NA) - Transcr Unit
TU1331 (NA) - Transcr Unit
TU1343 (NA) - Transcr Unit
TU1351 (NA) - Transcr Unit
TU1365 (NA) - Transcr Unit
TU1370 (NA) - Transcr Unit
TU1371 (NA) - Transcr Unit
TU138 (NA) - Transcr Unit
TU1382 (NA) - Transcr Unit
TU1387 (NA) - Transcr Unit
TU1401 (NA) - Transcr Unit
TU1419 (NA) - Transcr Unit
TU143 (NA) - Transcr Unit
TU1439 (NA) - Transcr Unit
TU1447 (NA) - Transcr Unit
TU1461 (NA) - Transcr Unit
TU1462 (NA) - Transcr Unit
TU1468 (NA) - Transcr Unit
TU147 (NA) - Transcr Unit
TU1475 (NA) - Transcr Unit
TU1481 (NA) - Transcr Unit
TU1483 (NA) - Transcr Unit
TU1484 (NA) - Transcr Unit
TU1514 (NA) - Transcr Unit
TU1527 (NA) - Transcr Unit
TU1528 (NA) - Transcr Unit
TU1535 (NA) - Transcr Unit
TU1536 (NA) - Transcr Unit
TU1539 (NA) - Transcr Unit
TU1544 (NA) - Transcr Unit
TU1547 (NA) - Transcr Unit
TU1548 (NA) - Transcr Unit
TU1550 (NA) - Transcr Unit
TU1557 (NA) - Transcr Unit
TU1558 (NA) - Transcr Unit
TU1586 (NA) - Transcr Unit
TU1588 (NA) - Transcr Unit
TU1609 (NA) - Transcr Unit
TU1612 (NA) - Transcr Unit
TU1639 (NA) - Transcr Unit
TU1641 (NA) - Transcr Unit
TU1652 (NA) - Transcr Unit
TU1671 (NA) - Transcr Unit
TU1678 (NA) - Transcr Unit
TU168 (NA) - Transcr Unit
TU1687 (NA) - Transcr Unit
TU1692 (NA) - Transcr Unit
TU1700 (NA) - Transcr Unit
TU171 (NA) - Transcr Unit
TU1721 (NA) - Transcr Unit
TU1724 (NA) - Transcr Unit
TU1729 (NA) - Transcr Unit
TU1751 (NA) - Transcr Unit
TU1755 (NA) - Transcr Unit
TU1764 (NA) - Transcr Unit
TU177 (NA) - Transcr Unit
TU1784 (NA) - Transcr Unit
TU1787 (NA) - Transcr Unit
TU1806 (NA) - Transcr Unit
TU182 (NA) - Transcr Unit
TU1826 (NA) - Transcr Unit
TU1841 (NA) - Transcr Unit
TU1846 (NA) - Transcr Unit
TU1850 (NA) - Transcr Unit
TU1877 (NA) - Transcr Unit
TU1896 (NA) - Transcr Unit
TU1897 (NA) - Transcr Unit
TU190 (NA) - Transcr Unit
TU1902 (NA) - Transcr Unit
TU1917 (NA) - Transcr Unit
TU1919 (NA) - Transcr Unit
TU1930 (NA) - Transcr Unit
TU1935 (NA) - Transcr Unit
TU1947 (NA) - Transcr Unit
TU1955 (NA) - Transcr Unit
TU1963 (NA) - Transcr Unit
TU197 (NA) - Transcr Unit
TU1981 (NA) - Transcr Unit
TU1987 (NA) - Transcr Unit
TU1988 (NA) - Transcr Unit
TU1996 (NA) - Transcr Unit
TU2004 (NA) - Transcr Unit
TU2037 (NA) - Transcr Unit
TU2051 (NA) - Transcr Unit
TU2059 (NA) - Transcr Unit
TU207 (NA) - Transcr Unit
TU208 (NA) - Transcr Unit
TU210 (NA) - Transcr Unit
TU2104 (NA) - Transcr Unit
TU2110 (NA) - Transcr Unit
TU2114 (NA) - Transcr Unit
TU2116 (NA) - Transcr Unit
TU2126 (NA) - Transcr Unit
TU2131 (NA) - Transcr Unit
TU2145 (NA) - Transcr Unit
TU2149 (NA) - Transcr Unit
TU2160 (NA) - Transcr Unit
TU2170 (NA) - Transcr Unit
TU2236 (NA) - Transcr Unit
TU2239 (NA) - Transcr Unit
TU226 (NA) - Transcr Unit
TU2261 (NA) - Transcr Unit
TU2262 (NA) - Transcr Unit
TU2263 (NA) - Transcr Unit
TU2266 (NA) - Transcr Unit
TU2268 (NA) - Transcr Unit
TU227 (NA) - Transcr Unit
TU2278 (NA) - Transcr Unit
TU2314 (NA) - Transcr Unit
TU2318 (NA) - Transcr Unit
TU2339 (NA) - Transcr Unit
TU2344 (NA) - Transcr Unit
TU2353 (NA) - Transcr Unit
TU2354 (NA) - Transcr Unit
TU2355 (NA) - Transcr Unit
TU2360 (NA) - Transcr Unit
TU2362 (NA) - Transcr Unit
TU2376 (NA) - Transcr Unit
TU239 (NA) - Transcr Unit
TU2407 (NA) - Transcr Unit
TU2413 (NA) - Transcr Unit
TU242 (NA) - Transcr Unit
TU2420 (NA) - Transcr Unit
TU2434 (NA) - Transcr Unit
TU2442 (NA) - Transcr Unit
TU2454 (NA) - Transcr Unit
TU2459 (NA) - Transcr Unit
TU2460 (NA) - Transcr Unit
TU2470 (NA) - Transcr Unit
TU2471 (NA) - Transcr Unit
TU2494 (NA) - Transcr Unit
TU2498 (NA) - Transcr Unit
TU2509 (NA) - Transcr Unit
TU2519 (NA) - Transcr Unit
TU252 (NA) - Transcr Unit
TU2520 (NA) - Transcr Unit
TU2560 (NA) - Transcr Unit
TU2579 (NA) - Transcr Unit
TU2605 (NA) - Transcr Unit
TU2614 (NA) - Transcr Unit
TU2616 (NA) - Transcr Unit
TU2627 (NA) - Transcr Unit
TU2630 (NA) - Transcr Unit
TU2645 (NA) - Transcr Unit
TU265 (NA) - Transcr Unit
TU2655 (NA) - Transcr Unit
TU2677 (NA) - Transcr Unit
TU2685 (NA) - Transcr Unit
TU2747 (NA) - Transcr Unit
TU2748 (NA) - Transcr Unit
TU2757 (NA) - Transcr Unit
TU2759 (NA) - Transcr Unit
TU276 (NA) - Transcr Unit
TU2762 (NA) - Transcr Unit
TU2763 (NA) - Transcr Unit
TU2788 (NA) - Transcr Unit
TU2791 (NA) - Transcr Unit
TU2814 (NA) - Transcr Unit
TU2827 (NA) - Transcr Unit
TU2837 (NA) - Transcr Unit
TU2839 (NA) - Transcr Unit
TU2848 (NA) - Transcr Unit
TU2861 (NA) - Transcr Unit
TU2868 (NA) - Transcr Unit
TU2869 (NA) - Transcr Unit
TU287 (NA) - Transcr Unit
TU2872 (NA) - Transcr Unit
TU2877 (NA) - Transcr Unit
TU2882 (NA) - Transcr Unit
TU2889 (NA) - Transcr Unit
TU2890 (NA) - Transcr Unit
TU2901 (NA) - Transcr Unit
TU2930 (NA) - Transcr Unit
TU2933 (NA) - Transcr Unit
TU2938 (NA) - Transcr Unit
TU2939 (NA) - Transcr Unit
TU294 (NA) - Transcr Unit
TU2973 (NA) - Transcr Unit
TU2974 (NA) - Transcr Unit
TU2978 (NA) - Transcr Unit
TU298 (NA) - Transcr Unit
TU2987 (NA) - Transcr Unit
TU3003 (NA) - Transcr Unit
TU3052 (NA) - Transcr Unit
TU3098 (NA) - Transcr Unit
TU3120 (NA) - Transcr Unit
TU3127 (NA) - Transcr Unit
TU313 (NA) - Transcr Unit
TU3131 (NA) - Transcr Unit
TU3139 (NA) - Transcr Unit
TU3178 (NA) - Transcr Unit
TU318 (NA) - Transcr Unit
TU3184 (NA) - Transcr Unit
TU319 (NA) - Transcr Unit
TU3204 (NA) - Transcr Unit
TU321 (NA) - Transcr Unit
TU3213 (NA) - Transcr Unit
TU3216 (NA) - Transcr Unit
TU3223 (NA) - Transcr Unit
TU3235 (NA) - Transcr Unit
TU3251 (NA) - Transcr Unit
TU3298 (NA) - Transcr Unit
TU3309 (NA) - Transcr Unit
TU3313 (NA) - Transcr Unit
TU3319 (NA) - Transcr Unit
TU3325 (NA) - Transcr Unit
TU3335 (NA) - Transcr Unit
TU3337 (NA) - Transcr Unit
TU3341 (NA) - Transcr Unit
TU3366 (NA) - Transcr Unit
TU3367 (NA) - Transcr Unit
TU3372 (NA) - Transcr Unit
TU3423 (NA) - Transcr Unit
TU3426 (NA) - Transcr Unit
TU3432 (NA) - Transcr Unit
TU3436 (NA) - Transcr Unit
TU345 (NA) - Transcr Unit
TU3456 (NA) - Transcr Unit
TU3468 (NA) - Transcr Unit
TU3475 (NA) - Transcr Unit
TU35 (NA) - Transcr Unit
TU3505 (NA) - Transcr Unit
TU3511 (NA) - Transcr Unit
TU3515 (NA) - Transcr Unit
TU3573 (NA) - Transcr Unit
TU3576 (NA) - Transcr Unit
TU3577 (NA) - Transcr Unit
TU359 (NA) - Transcr Unit
TU3598 (NA) - Transcr Unit
TU360 (NA) - Transcr Unit
TU361 (NA) - Transcr Unit
TU3616 (NA) - Transcr Unit
TU3618 (NA) - Transcr Unit
TU3655 (NA) - Transcr Unit
TU3659 (NA) - Transcr Unit
TU3660 (NA) - Transcr Unit
TU3664 (NA) - Transcr Unit
TU368 (NA) - Transcr Unit
TU3696 (NA) - Transcr Unit
TU3712 (NA) - Transcr Unit
TU372 (NA) - Transcr Unit
TU376 (NA) - Transcr Unit
TU381 (NA) - Transcr Unit
TU389 (NA) - Transcr Unit
TU390 (NA) - Transcr Unit
TU392 (NA) - Transcr Unit
TU396 (NA) - Transcr Unit
TU397 (NA) - Transcr Unit
TU41 (NA) - Transcr Unit
TU412 (NA) - Transcr Unit
TU414 (NA) - Transcr Unit
TU419 (NA) - Transcr Unit
TU421 (NA) - Transcr Unit
TU425 (NA) - Transcr Unit
TU43 (NA) - Transcr Unit
TU440 (NA) - Transcr Unit
TU45 (NA) - Transcr Unit
TU458 (NA) - Transcr Unit
TU468 (NA) - Transcr Unit
TU472 (NA) - Transcr Unit
TU474 (NA) - Transcr Unit
TU48 (NA) - Transcr Unit
TU480 (NA) - Transcr Unit
TU482 (NA) - Transcr Unit
TU499 (NA) - Transcr Unit
TU5004 (NA) - Transcr Unit
TU5008 (NA) - Transcr Unit
TU5013 (NA) - Transcr Unit
TU5015 (NA) - Transcr Unit
TU5020 (NA) - Transcr Unit
TU5027 (NA) - Transcr Unit
TU5028 (NA) - Transcr Unit
TU5050 (NA) - Transcr Unit
TU5061 (NA) - Transcr Unit
TU5066 (NA) - Transcr Unit
TU5078 (NA) - Transcr Unit
TU5083 (NA) - Transcr Unit
TU5112 (NA) - Transcr Unit
TU5116 (NA) - Transcr Unit
TU540 (NA) - Transcr Unit
TU551 (NA) - Transcr Unit
TU563 (NA) - Transcr Unit
TU566 (NA) - Transcr Unit
TU575 (NA) - Transcr Unit
TU58 (NA) - Transcr Unit
TU583 (NA) - Transcr Unit
TU594 (NA) - Transcr Unit
TU6 (NA) - Transcr Unit
TU6003 (NA) - Transcr Unit
TU6004 (NA) - Transcr Unit
TU6008 (NA) - Transcr Unit
TU6014 (NA) - Transcr Unit
TU6019 (NA) - Transcr Unit
TU6026 (NA) - Transcr Unit
TU6028 (NA) - Transcr Unit
TU6032 (NA) - Transcr Unit
TU6051 (NA) - Transcr Unit
TU6059 (NA) - Transcr Unit
TU6063 (NA) - Transcr Unit
TU6069 (NA) - Transcr Unit
TU607 (NA) - Transcr Unit
TU6074 (NA) - Transcr Unit
TU6080 (NA) - Transcr Unit
TU6083 (NA) - Transcr Unit
TU6097 (NA) - Transcr Unit
TU62 (NA) - Transcr Unit
TU639 (NA) - Transcr Unit
TU646 (NA) - Transcr Unit
TU648 (NA) - Transcr Unit
TU652 (NA) - Transcr Unit
TU658 (NA) - Transcr Unit
TU684 (NA) - Transcr Unit
TU686 (NA) - Transcr Unit
TU699 (NA) - Transcr Unit
TU7 (NA) - Transcr Unit
TU7005 (NA) - Transcr Unit
TU7009 (NA) - Transcr Unit
TU7010 (NA) - Transcr Unit
TU7012 (NA) - Transcr Unit
TU7031 (NA) - Transcr Unit
TU7039 (NA) - Transcr Unit
TU7040 (NA) - Transcr Unit
TU7041 (NA) - Transcr Unit
TU7045 (NA) - Transcr Unit
TU7048 (NA) - Transcr Unit
TU705 (NA) - Transcr Unit
TU7050 (NA) - Transcr Unit
TU7056 (NA) - Transcr Unit
TU7070 (NA) - Transcr Unit
TU7073 (NA) - Transcr Unit
TU708 (NA) - Transcr Unit
TU7081 (NA) - Transcr Unit
TU7085 (NA) - Transcr Unit
TU7086 (NA) - Transcr Unit
TU7090 (NA) - Transcr Unit
TU7093 (NA) - Transcr Unit
TU711 (NA) - Transcr Unit
TU714 (NA) - Transcr Unit
TU723 (NA) - Transcr Unit
TU726 (NA) - Transcr Unit
TU727 (NA) - Transcr Unit
TU728 (NA) - Transcr Unit
TU734 (NA) - Transcr Unit
TU735 (NA) - Transcr Unit
TU743 (NA) - Transcr Unit
TU744 (NA) - Transcr Unit
TU746 (NA) - Transcr Unit
TU760 (NA) - Transcr Unit
TU782 (NA) - Transcr Unit
TU8004 (NA) - Transcr Unit
TU8017 (NA) - Transcr Unit
TU8020 (NA) - Transcr Unit
TU8029 (NA) - Transcr Unit
TU8038 (NA) - Transcr Unit
TU8039 (NA) - Transcr Unit
TU8045 (NA) - Transcr Unit
TU811 (NA) - Transcr Unit
TU822 (NA) - Transcr Unit
TU824 (NA) - Transcr Unit
TU825 (NA) - Transcr Unit
TU838 (NA) - Transcr Unit
TU840 (NA) - Transcr Unit
TU843 (NA) - Transcr Unit
TU846 (NA) - Transcr Unit
TU847 (NA) - Transcr Unit
TU85 (NA) - Transcr Unit
TU855 (NA) - Transcr Unit
TU86 (NA) - Transcr Unit
TU861 (NA) - Transcr Unit
TU868 (NA) - Transcr Unit
TU871 (NA) - Transcr Unit
TU872 (NA) - Transcr Unit
TU873 (NA) - Transcr Unit
TU883 (NA) - Transcr Unit
TU884 (NA) - Transcr Unit
TU887 (NA) - Transcr Unit
TU892 (NA) - Transcr Unit
TU894 (NA) - Transcr Unit
TU900 (NA) - Transcr Unit
TU902 (NA) - Transcr Unit
TU903 (NA) - Transcr Unit
TU906 (NA) - Transcr Unit
TU913 (NA) - Transcr Unit
TU92 (NA) - Transcr Unit
TU932 (NA) - Transcr Unit
TU952 (NA) - Transcr Unit
TU953 (NA) - Transcr Unit
TU955 (NA) - Transcr Unit
TU960 (NA) - Transcr Unit
TU961 (NA) - Transcr Unit
TU973 (NA) - Transcr Unit
TU980 (NA) - Transcr Unit
TU992 (NA) - Transcr Unit
nadA/sll0622 (mRNA / CDS) - TU3528
nadA/sll0622 (Protein) - TU3528
nadB/sll0631 (mRNA / CDS) - TU3500
nadB/sll0631 (Protein) - TU3500
TU3500 (sll0631) - Transcr Unit
nadC/slr0936 (mRNA / CDS) - TU2072
nadC/slr0936 (Protein) - TU2072
nadE/slr1691 (mRNA / CDS) - TU2058
nadE/slr1691 (Protein) - TU2058
napC/sll0873 (mRNA / CDS)
apcE/slr0335 (Protein) - TU2385
narB/sll1454 (mRNA / CDS) - TU1023
narB/sll1454 (Protein) - TU1023
natA/slr0467 (Protein) - TU2739
TU2739 (slr0467, slr0468) - Transcr Unit
natB/slr0559 (mRNA / CDS) - TU2947
natB/slr0559 (Protein) - TU2947
TU2947 (slr0559) - Transcr Unit
natC/sll0146 (mRNA / CDS) - TU2276
TU2276 (sll0146) - Transcr Unit
natC/sll0146,ssl3127 (Protein)
livH/slr0949 (mRNA / CDS) - TU2088
natD/slr0949 (Protein) - TU2088
TU2088 (slr0949) - Transcr Unit
natE/slr1881 (mRNA / CDS) - TU1269
natE/slr1881 (Protein) - TU1269
TU1562 (ssl0452, ssl0453) - Transcr Unit
nbp1,rps1b/slr1984 (mRNA / CDS) - TU1878
nbp1,rps1b/slr1984 (Protein) - TU1878
TU1878 (slr1984) - Transcr Unit
TU123 (ncl0040) - Transcr Unit
TU194 (ncl0050) - Transcr Unit
TU201 (ncl0060, sll1033) - Transcr Unit
TU205 (ncl0070, sll1028, sll1029, sll1030, sll1031, sll1032) - Transcr Unit
TU331 (ncl0110, sll0934, ncl0100) - Transcr Unit
TU463 (ncl0150, sll1503, sll1504, sll1505) - Transcr Unit
TU475 (ncl0160) - Transcr Unit
TU491 (ncl0170, sll1009) - Transcr Unit
TU581 (ncl0215) - Transcr Unit
TU670 (ncl0240) - Transcr Unit
TU715 (ncl0270) - Transcr Unit
TU766 (ncl0310, sll1921) - Transcr Unit
TU784 (ncl0320, sll1910) - Transcr Unit
TU798 (ncl0350) - Transcr Unit
TU795 (ncl0350, sll1077, sll1078, sll1079) - Transcr Unit
TU823 (ncl0360, ssl2084, sll1069) - Transcr Unit
TU830 (ncl0370, ssl3446, sll1823, sll1824) - Transcr Unit
TU837 (ncl0380, sll1799, sll1800, sll1801, sll1802, ssl3432, sll1803, sll1804, sll1805, ssl3436, ssl3437, sll1806, sll1807, sll1808, sll1809, sll1810, sll1811, sll1812, sll1813) - Transcr Unit
TU853 (ncl0390) - Transcr Unit
TU908 (ncl0400, sml0012, sll1112) - Transcr Unit
TU915 (ncl0410, sll1967) - Transcr Unit
TU938 (ncl0420, sll1739) - Transcr Unit
TU1146 (ncl0490) - Transcr Unit
TU1144 (ncl0490, ssl2501, ssl2502) - Transcr Unit
TU1186 (ncl0500) - Transcr Unit
TU1185 (ncl0500, sll0871) - Transcr Unit
TU1288 (ncl0530, sll2012, sll2013, sll2014) - Transcr Unit
TU1338 (ncl0560, sll1689) - Transcr Unit
TU1341 (ncl0570, sll1687) - Transcr Unit
TU1431 (ncl0600, sll1151) - Transcr Unit
TU1440 (ncl0620) - Transcr Unit
TU1470 (ncl0640) - Transcr Unit
TU1494 (ncl0650, sll1613) - Transcr Unit
TU1497 (ncl0660, sll1608, sll1609) - Transcr Unit
TU1521 (ncl0690, sll1594) - Transcr Unit
TU1593 (ncl0710, ssl3769) - Transcr Unit
TU1658 (ncl0780) - Transcr Unit
TU1684 (ncl0790, sll1239, sll1240, sll1241) - Transcr Unit
TU1714 (ncl0800, sll1220, sll1221, sll1222, sll1223, sll1224, ssl2420, sll1225, sll1226) - Transcr Unit
TU1716 (ncl0810) - Transcr Unit
TU1737 (ncl0820, sll0815, sll0816, sll0817) - Transcr Unit
TU1825 (ncl0850, sll1895) - Transcr Unit
TU1829 (ncl0860, sll1894) - Transcr Unit
TU1931 (ncl0910) - Transcr Unit
TU1997 (ncl0950) - Transcr Unit
TU2054 (ncl0980) - Transcr Unit
TU2125 (ncl1010, sll1533, sll1534) - Transcr Unit
TU2142 (ncl1020) - Transcr Unit
TU2166 (ncl1040, sll0427) - Transcr Unit
TU2191 (ncl1045, sll0267, sll0268, sll0269, sll0270) - Transcr Unit
TU2206 (ncl1060) - Transcr Unit
TU2340 (ncl1090) - Transcr Unit
TU2356 (ncl1120, sll0947, ncl1110) - Transcr Unit
TU2366 (ncl1130) - Transcr Unit
TU2406 (ncl1150, sll0169) - Transcr Unit
TU2411 (ncl1160, sll0163, sll0166) - Transcr Unit
TU2474 (ncl1180, sll0330) - Transcr Unit
TU2507 (ncl1210, sll0764) - Transcr Unit
TU2518 (ncl1220, sll0750) - Transcr Unit
TU2646 (ncl1260, sll0199) - Transcr Unit
TU2824 (ncl1330) - Transcr Unit
TU2857 (ncl1350) - Transcr Unit
TU2884 (ncl1390, sll0185) - Transcr Unit
TU2924 (ncl1400) - Transcr Unit
TU3007 (ncl1450) - Transcr Unit
TU3022 (ncl1460, sll0821) - Transcr Unit
TU3100 (ncl1470, sll0470, sll0471) - Transcr Unit
TU3138 (ncl1500, sll0108, ncl1490) - Transcr Unit
TU3195 (ncl1530, sll0283) - Transcr Unit
TU3315 (ncl1590) - Transcr Unit
TU3314 (ncl1590, sll0038) - Transcr Unit
TU3378 (ncl1620, sll0504) - Transcr Unit
TU3410 (ncl1630, sll0895, sll0896) - Transcr Unit
TU3414 (ncl1650, sll0534, sll0535) - Transcr Unit
TU3503 (ncl1690, sll0629) - Transcr Unit
TU3523 (ncl1710, sll0624, sll0625, sll0626) - Transcr Unit
TU3536 (ncl1720) - Transcr Unit
TU3718 (ncl1800, SyR52) - Transcr Unit
TU87 (ncr0020, slr1129, slr1130) - Transcr Unit
TU105 (ncr0050, slr0728) - Transcr Unit
TU110 (ncr0060, slr0733) - Transcr Unit
TU127 (ncr0070, slr0242, slr0243) - Transcr Unit
TU162 (ncr0080, slr0257) - Transcr Unit
TU206 (ncr0100) - Transcr Unit
TU341 (ncr0150, slr0974) - Transcr Unit
TU409 (ncr0190, slr0687) - Transcr Unit
TU420 (ncr0200) - Transcr Unit
TU422 (ncr0200, ssr1155, slr0689) - Transcr Unit
TU433 (ncr0210) - Transcr Unit
TU434 (ncr0210) - Transcr Unit
TU443 (ncr0220, slr1597) - Transcr Unit
TU528 (ncr0240, slr1770, slr1771) - Transcr Unit
TU611 (ncr0270, slr1915) - Transcr Unit
TU643 (ncr0280, slr1028) - Transcr Unit
TU731 (ncr0310) - Transcr Unit
TU814 (ncr0350, slr1149, slr1150) - Transcr Unit
TU994 (ncr0400, slr1537, slr1538) - Transcr Unit
TU1041 (ncr0420, slr1220, slr1222) - Transcr Unit
TU1103 (ncr0440, slr1441) - Transcr Unit
TU1191 (ncr0470, slr0885) - Transcr Unit
TU1227 (ncr0480, slr1852, slr1853, slr1854, slr1855, slr1856, slr1857, slr1859, slr1860, slr1861) - Transcr Unit
TU1360 (ncr0510, slr1738) - Transcr Unit
TU1455 (ncr0540, slr2058, slr2059, slr2060) - Transcr Unit
TU1471 (ncr0570) - Transcr Unit
TU1628 (ncr0630, slr1505) - Transcr Unit
TU1695 (ncr0670) - Transcr Unit
TU1715 (ncr0700, ncr0710, ssr2227) - Transcr Unit
TU1748 (ncr0720) - Transcr Unit
TU1761 (ncr0730) - Transcr Unit
TU1847 (ncr0760, slr1971) - Transcr Unit
TU1852 (ncr0770, slr1972) - Transcr Unit
TU1953 (ncr0840) - Transcr Unit
TU1954 (ncr0840, slr1279, slr1280, slr1281) - Transcr Unit
TU1957 (ncr0860, slr1285) - Transcr Unit
TU2009 (ncr0890, slr1167, slr1168, slr1169) - Transcr Unit
TU2014 (ncr0900, slr1173, ssr1951) - Transcr Unit
TU2027 (ncr0920, slr1678) - Transcr Unit
TU2036 (ncr0930, slr1681, slr1682) - Transcr Unit
TU2098 (ncr0960, ssr1600, slr0954, slr0955) - Transcr Unit
TU2144 (ncr0970, slr1643) - Transcr Unit
TU2150 (ncr0980, slr1648) - Transcr Unit
TU2183 (ncr1010) - Transcr Unit
TU2184 (ncr1010, slr0447) - Transcr Unit
TU2224 (ncr1030, slr0397) - Transcr Unit
TU2296 (ncr1071) - Transcr Unit
TU2383 (ncr1100) - Transcr Unit
TU2440 (ncr1160) - Transcr Unit
TU2529 (ncr1220) - Transcr Unit
TU2555 (ncr1240, slr0355, slr0356) - Transcr Unit
TU2597 (ncr1280, slr0017) - Transcr Unit
TU2598 (ncr1280, slr0017) - Transcr Unit
TU2657 (ncr1320, slr0229) - Transcr Unit
TU2680 (ncr1330) - Transcr Unit
TU2746 (ncr1350, slr0473, slr0474) - Transcr Unit
TU2809 (ncr1360, smr0015) - Transcr Unit
TU2899 (ncr1420) - Transcr Unit
TU2969 (ncr1450, slr0579) - Transcr Unit
TU2988 (ncr1460, slr0829) - Transcr Unit
TU3039 (ncr1490, slr0079) - Transcr Unit
TU3149 (ncr1530, slr0105, slr0106) - Transcr Unit
TU3304 (ncr1560) - Transcr Unit
TU3330 (ncr1570, slr0038, slr0039) - Transcr Unit
TU3402 (ncr1580, slr0927) - Transcr Unit
TU3417 (ncr1590, slr0549, slr0550) - Transcr Unit
TU3419 (ncr1600, slr0551, slr0552, slr0553) - Transcr Unit
TU3458 (ncr1620) - Transcr Unit
TU3464 (ncr1630, slr1429) - Transcr Unit
TU3492 (ncr1640, slr1463) - Transcr Unit
TU3504 (ncr1650, slr0653) - Transcr Unit
TU3510 (ncr1660, slr0654, slr0655) - Transcr Unit
TU3599 (ncr1680, slr0753) - Transcr Unit
TU3604 (ncr1690) - Transcr Unit
TU3676 (ncr1710, slr0459) - Transcr Unit
TU3677 (ncr1720, slr0460) - Transcr Unit
TU1375 (slr0851) - Transcr Unit
ndbB/slr1743 (mRNA / CDS) - TU1367
ndbB/slr1743 (Protein) - TU1367
TU1367 (slr1743) - Transcr Unit
ndbC/sll1484 (mRNA / CDS) - TU3571
ndbC/sll1484 (Protein) - TU3571
ndhA/sll0519 (mRNA / CDS) - TU3434
ndhA/sll0519 (Protein) - TU3434
TU3434 (sll0519, sll0520, sll0521, sll0522) - Transcr Unit
ndhB/sll0223 (mRNA / CDS) - TU146
ndhB/sll0223 (Protein) - TU146
TU146 (sll0223) - Transcr Unit
sll0223-5UTR (5'UTR) - TU146
sll0223-as (asRNA)
ndhD1/slr0331 (mRNA / CDS) - TU2377
ndhD1/slr0331 (Protein) - TU2377
TU2377 (slr0331) - Transcr Unit
TU282 (slr1291) - Transcr Unit
ndhD3/sll1733 (mRNA / CDS) - TU951
ndhD3/sll1733 (Protein) - TU951
ndhD4/sll0027 (mRNA / CDS) - TU3338
ndhD4/sll0027 (Protein) - TU3338
sll0027-as (asRNA)
ndhD5/slr2007 (mRNA / CDS) - TU756
ndhD5/slr2007 (Protein) - TU756
ndhD6/slr2009 (mRNA / CDS) - TU757
ndhD6/slr2009 (Protein) - TU757
ndhE/sll0522 (mRNA / CDS) - TU3434
ndhE/sll0522 (Protein) - TU3434
ndhF1/slr0844 (mRNA / CDS) - TU3013
ndhF1/slr0844 (Protein) - TU3013
TU3013 (slr0844) - Transcr Unit
slr0844-3UTR (3'UTR) - TU3013
slr0844-5UTR (5'UTR) - TU3013
ndhF3/sll1732 (mRNA / CDS) - TU951
ndhF3/sll1732 (Protein) - TU951
sll1732-5UTR (5'UTR) - TU951
TU951 (sll1732, sll1733, sll1734) - Transcr Unit
ndhF4/sll0026 (mRNA / CDS) - TU3338
ndhF4/sll0026 (Protein) - TU3338
TU3338 (sll0026, sll0027) - Transcr Unit
ndhG/sll0521 (mRNA / CDS) - TU3434
ndhG/sll0521 (Protein) - TU3434
ndhH/slr0261 (mRNA / CDS) - TU1546
ndhH/slr0261 (Protein) - TU1546
slr0261-as (asRNA)
slr0261-as2 (asRNA)
slr0261-as3 (asRNA)
TU1546 (slr0261, slr0262, slr0263, slr0264) - Transcr Unit
ndhI/sll0520 (mRNA / CDS) - TU3434
ndhI/sll0520 (Protein) - TU3434
ndhJ/slr1281 (mRNA / CDS) - TU1954
ndhJ/slr1281 (Protein) - TU1954
ndhK/slr1280 (mRNA / CDS) - TU1954
ndhK/slr1280 (Protein) - TU1954
ndhL/ssr1386 (mRNA / CDS) - TU1749
ndhL/ssr1386 (Protein) - TU1749
TU1749 (ssr1386, slr0815, slr0816) - Transcr Unit
ndhM/slr1623 (mRNA / CDS) - TU2113
ndhM/slr1623 (Protein) - TU2113
ndhN/sll1262 (mRNA / CDS) - TU1776
ndhN/sll1262 (Protein) - TU1776
ndhO/ssl1690 (mRNA / CDS) - TU2940
ndhO/ssl1690 (Protein) - TU2940
rbcR/sll1594 (mRNA / CDS) - TU1521
ndhR/sll1594 (Protein) - TU1521
ndkR/sll1852 (mRNA / CDS) - TU2319
ndkR/sll1852 (Protein) - TU2319
nhaS1/slr1727 (mRNA / CDS) - TU1524
nhaS1/slr1727 (Protein) - TU1524
nhaS2/sll0273 (mRNA / CDS) - TU2187
nhaS2/sll0273 (Protein) - TU2187
nhaS3/sll0689 (mRNA / CDS) - TU2780
nhaS3/sll0689 (Protein) - TU2780
TU2780 (sll0689) - Transcr Unit
nhaS5/slr0415 (mRNA / CDS) - TU2833
nhaS5/slr0415 (Protein) - TU2833
TU2833 (slr0415) - Transcr Unit
slr0415-as3 (asRNA)
nifJ/sll0741 (mRNA / CDS) - TU3296
nifJ/sll0741 (Protein) - TU3296
sll0741-5UTR (5'UTR) - TU3296
TU108 (sll0704) - Transcr Unit
nifS/slr0077 (mRNA / CDS) - TU3030
nifS/slr0077 (Protein) - TU3030
nifS/slr0387 (mRNA / CDS) - TU2487
nifS/slr0387 (Protein) - TU2487
TU2487 (slr0387, slr0388) - Transcr Unit
nirA/slr0898 (mRNA / CDS) - TU2925
nirA/slr0898 (Protein) - TU2925
TU2925 (slr0898, slr0899, slr0900, slr0901, slr0902, ssr1527, slr0903) - Transcr Unit
nlpD/slr0993 (mRNA / CDS) - TU2352
nlpD/slr0993 (Protein) - TU2352
slr0993-as (asRNA)
slr0993-as2 (asRNA)
nnr/sll1433 (mRNA / CDS) - TU1978
nnr/sll1433 (Protein) - TU1978
norB/sll0450 (mRNA / CDS)
norB/sll0450 (Protein)
Norf1 (mRNA / CDS) - TU292
Norf1 (Protein) - TU292
Norf1-5UTR (5'UTR) - TU292
TU292 (Norf1, sll1193) - Transcr Unit
Norf2 (mRNA / CDS) - TU355
Norf2 (Protein) - TU355
TU1188 (Norf4) - Transcr Unit
TU1457 (Norf5) - Transcr Unit
TU1918 (Norf6, sll1374) - Transcr Unit
Norf7 (mRNA / CDS) - TU2506
Norf7 (Protein) - TU2506
TU2506 (Norf7) - Transcr Unit
Norf7-3UTR (3'UTR) - TU2506
nplT/sll0842 (mRNA / CDS) - TU1402
nplT/sll0842 (Protein) - TU1402
sll0842-5UTR (5'UTR) - TU1402
nrdA/slr1164 (mRNA / CDS) - TU2003
nrdA/slr1164 (Protein) - TU2003
slr1164-as (asRNA)
nrdR/slr0780 (mRNA / CDS) - TU2513
nrdR/slr0780 (Protein) - TU2513
slr0780-5UTR (5'UTR) - TU2513
TU3220 (slr0796) - Transcr Unit
nrtA/sll1450 (mRNA / CDS) - TU1023
nrtA/sll1450 (Protein) - TU1023
sll1450-5UTR (5'UTR) - TU1023
TU1023 (sll1450, sll1451, sll1452, sll1453, ssl2814, sll1454) - Transcr Unit
nrtB/sll1451 (mRNA / CDS) - TU1023
nrtB/sll1451 (Protein) - TU1023
nrtC/sll1452 (mRNA / CDS) - TU1023
nrtC/sll1452 (Protein) - TU1023
nrtD/sll1082 (mRNA / CDS)
nrtD/sll1082 (Protein)
nrtD/sll1453 (mRNA / CDS) - TU1023
nrtD/sll1453 (Protein) - TU1023
nsiR4/ncl0540 (ncRNA) - TU1322
TU1322 (nsiR4, sll1698, sll1699) - Transcr Unit
ntcA, ycf28/sll1423 (mRNA / CDS) - TU1630
ntcA, ycf28/sll1423 (Protein) - TU1630
TU1630 (sll1423, sll1424) - Transcr Unit
TU2218 (slr0395) - Transcr Unit
nucH/sll0656 (mRNA / CDS) - TU388
nucH/sll0656 (Protein) - TU388
nusA/slr0743 (mRNA / CDS) - TU119
nusA/slr0743 (Protein) - TU119
nusB/sll0271 (mRNA / CDS) - TU2188
nusB/sll0271 (Protein) - TU2188
nusG/sll1742 (mRNA / CDS) - TU931
nusG/sll1742 (Protein) - TU931
nylA/sll0828 (mRNA / CDS) - TU3005
nylA/sll0828 (Protein) - TU3005
obgE/slr1090 (mRNA / CDS) - TU379
obgE/slr1090 (Protein) - TU379
odhB/sll1841 (mRNA / CDS) - TU2343
odhB/sll1841 (Protein) - TU2343
sll1841-3UTR (3'UTR) - TU2343
opcA/slr1734 (mRNA / CDS) - TU1349
opcA/slr1734 (Protein) - TU1349
TU1349 (slr1734) - Transcr Unit
pacL/sll0672 (mRNA / CDS) - TU3268
pacL/sll0672 (Protein) - TU3268
TU3268 (sll0672) - Transcr Unit
sll0672-3UTR (3'UTR) - TU3268
pacL/sll1076 (mRNA / CDS) - TU802
pacL/sll1076 (Protein) - TU802
pacS/sll1920 (mRNA / CDS) - TU767
pacS/sll1920 (Protein) - TU767
TU767 (sll1920) - Transcr Unit
TU768 (sll1920) - Transcr Unit
panB/slr0526 (mRNA / CDS) - TU3346
panB/slr0526 (Protein) - TU3346
panC/sll1249 (mRNA / CDS) - TU1798
panC/sll1249 (Protein) - TU1798
TU1798 (sll1249, sll1250) - Transcr Unit
panD/sll0892 (mRNA / CDS) - TU2940
panD/sll0892 (Protein) - TU2940
PBP/sll1167 (mRNA / CDS) - TU1959
PBP/sll1167 (Protein) - TU1959
pcnB/sll1253 (mRNA / CDS) - TU1794
pcnB/sll1253 (Protein) - TU1794
pcrA/sll1143 (mRNA / CDS) - TU1043
pcrA/sll1143 (Protein) - TU1043
TU1043 (sll1143) - Transcr Unit
pcyA/slr0116 (mRNA / CDS) - TU3162
pcyA/slr0116 (Protein) - TU3162
pdhA/slr1934 (mRNA / CDS) - TU2316
pdhA/slr1934 (Protein) - TU2316
pdhB/sll1721 (mRNA / CDS) - TU966
pdhB/sll1721 (Protein) - TU966
sll1721-5UTR (5'UTR) - TU966
pds, crtD, crtP/slr1254 (mRNA / CDS) - TU1438
pds, crtD, crtP/slr1254 (Protein) - TU1438
slr1254-as (asRNA)
TU1438 (slr1254, slr1255) - Transcr Unit
pdxA/sll0660 (mRNA / CDS) - TU3287
pdxA/sll0660 (Protein) - TU3287
TU3287 (sll0660) - Transcr Unit
pdxH/sll1440 (mRNA / CDS) - TU1968
pdxH/sll1440 (Protein) - TU1968
TU1968 (sll1440) - Transcr Unit
pdxJ/slr1779 (mRNA / CDS) - TU224
pdxJ/slr1779 (Protein) - TU224
TU224 (slr1779, slr1780) - Transcr Unit
pepP/sll0136 (mRNA / CDS) - TU2286
pepP/sll0136 (Protein) - TU2286
petA/sll1317 (mRNA / CDS) - TU1200
petA/sll1317 (Protein) - TU1200
petB/slr0342 (mRNA / CDS) - TU2526
petB/slr0342 (Protein) - TU2526
TU2526 (slr0342) - Transcr Unit
slr0342-3UTR (3'UTR) - TU2526
slr0342-5UTR (5'UTR) - TU2526
petC1/sll1316 (mRNA / CDS) - TU1200
petC1/sll1316 (Protein) - TU1200
sll1316-5UTR (5'UTR) - TU1200
TU1200 (sll1316, sll1317, sll0860, sll0861, sll0862, sll0863) - Transcr Unit
petC/slr1185 (mRNA / CDS)
petC2/slr1185 (Protein)
petC3/sll1182 (mRNA / CDS) - TU1926
petC3/sll1182 (Protein) - TU1926
sll1182-5UTR (5'UTR) - TU1926
TU1926 (sll1182, sll1369, sll1370) - Transcr Unit
petD/slr0343 (mRNA / CDS) - TU2528
petD/slr0343 (Protein) - TU2528
TU2528 (slr0343) - Transcr Unit
slr0343-3UTR (3'UTR) - TU2528
slr0343-5UTR (5'UTR) - TU2528
petE/sll0199 (mRNA / CDS) - TU2646
petE/sll0199 (Protein) - TU2646
petF/ssl0020 (mRNA / CDS) - TU2593
petF/ssl0020 (Protein) - TU2593
TU2593 (ssl0020) - Transcr Unit
ssl0020-3UTR (3'UTR) - TU2593
ssl0020-5UTR (5'UTR) - TU2593
petF, fdx/sll1382 (mRNA / CDS) - TU1903
petF, fdx/sll1382 (Protein) - TU1903
sll1382-5UTR (5'UTR) - TU1903
petF, fdx/slr0150 (mRNA / CDS) - TU2264
petF, fdx/slr0150 (Protein) - TU2264
petF, fdx/slr1828 (Protein) - TU944
TU944 (slr1828) - Transcr Unit
TU1903 (sll1382, sll1383) - Transcr Unit
TU1891 (smr0010, ncr0800) - Transcr Unit
petH/slr1643 (mRNA / CDS) - TU2144
petH/slr1643 (Protein) - TU2144
petJ/sll1796 (mRNA / CDS)
petJ/sll1796 (Protein)
TU3289 (smr0003) - Transcr Unit
TU149 (sml0004, sgl0002) - Transcr Unit
TU150 (sml0004, sgl0002) - Transcr Unit
pfkA/sll0745 (mRNA / CDS) - TU3296
pfkA/sll0745 (Protein) - TU3296
pfkA/sll1196 (mRNA / CDS) - TU290
pfkA/sll1196 (Protein) - TU290
pgi/slr1349 (mRNA / CDS) - TU1802
pgi/slr1349 (Protein) - TU1802
pgk/slr0394 (mRNA / CDS) - TU2217
pgk/slr0394 (Protein) - TU2217
TU2217 (slr0394) - Transcr Unit
slr0394-3UTR (3'UTR) - TU2217
slr0394-as (asRNA)
pgm/sll0726 (mRNA / CDS) - TU3610
pgm/sll0726 (Protein) - TU3610
pgsA/sll1522 (mRNA / CDS) - TU2154
pgsA/sll1522 (Protein) - TU2154
sll1522-5UTR (5'UTR) - TU2154
phaA/slr1993 (mRNA / CDS) - TU1478
phaA/slr1993 (Protein) - TU1478
TU1478 (slr1993, slr1994) - Transcr Unit
phaB/slr1994 (mRNA / CDS) - TU1478
phaB/slr1994 (Protein) - TU1478
phaC/slr1830 (mRNA / CDS) - TU946
phaC/slr1830 (Protein) - TU946
phaE/slr1829 (mRNA / CDS) - TU946
phaE/slr1829 (Protein) - TU946
TU946 (slr1829, slr1830) - Transcr Unit
Phb/slr1106 (mRNA / CDS) - TU199
Phb/slr1106 (Protein) - TU199
phdD/slr1096 (mRNA / CDS) - TU186
phdD/slr1096 (Protein) - TU186
pheA/sll1662 (mRNA / CDS) - TU255
pheA/sll1662 (Protein) - TU255
pheS/sll0454 (mRNA / CDS) - TU2776
pheS/sll0454 (Protein) - TU2776
TU2776 (sll0454) - Transcr Unit
pheT/sll1553 (mRNA / CDS) - TU2065
pheT/sll1553 (Protein) - TU2065
TU2065 (sll1553) - Transcr Unit
phhB/ssl2296 (Protein) - TU1429
ssl2296-3UTR (3'UTR) - TU1429
phoR, hik7/sll0337 (mRNA / CDS) - TU2458
phoR, hik7/sll0337 (Protein) - TU2458
TU2458 (sll0337) - Transcr Unit
phoU/slr0741 (mRNA / CDS) - TU118
phoU/slr0741 (Protein) - TU118
phrA/slr0854 (mRNA / CDS) - TU1378
phrA/slr0854 (Protein) - TU1378
TU1378 (slr0854) - Transcr Unit
TU1379 (slr0854) - Transcr Unit
slr0854-as (asRNA)
pilA1/sll1694 (mRNA / CDS) - TU1330
pilA1/sll1694 (Protein) - TU1330
sll1694-5UTR (5'UTR) - TU1330
TU1330 (sll1694, sll1695, sll1696) - Transcr Unit
pilA10/slr2016 (mRNA / CDS) - TU763
pilA10/slr2016 (Protein) - TU763
pilA11/slr2017 (mRNA / CDS) - TU763
pilA11/slr2017 (Protein) - TU763
pilA2/sll1695 (mRNA / CDS) - TU1330
pilA2/sll1695 (Protein) - TU1330
pilA5/slr1928 (mRNA / CDS) - TU2300
pilA5/slr1928 (Protein) - TU2300
slr1928-5UTR (5'UTR) - TU2300
TU2300 (slr1928, slr1929) - Transcr Unit
pilA6/slr1929 (mRNA / CDS) - TU2300
pilA6/slr1929 (Protein) - TU2300
TU2301 (slr1930, slr1931, slr1932) - Transcr Unit
pilA8/slr1931 (mRNA / CDS) - TU2301
pilA8/slr1931 (Protein) - TU2301
pilA9/slr2015 (mRNA / CDS) - TU763
pilA9/slr2015 (Protein) - TU763
slr2015-5UTR (5'UTR) - TU763
TU763 (slr2015, slr2016, slr2017, slr2018, slr2019) - Transcr Unit
pilB1/slr0063 (mRNA / CDS) - TU2725
pilB1/slr0063 (Protein) - TU2725
slr0063-5UTR (5'UTR) - TU2725
TU2723 (slr0063, slr0064) - Transcr Unit
TU2725 (slr0063, slr0064) - Transcr Unit
pilC/slr0162 (mRNA / CDS) - TU2283
pilC/slr0162 (Protein) - TU2283
pilC'/slr0163 (mRNA / CDS) - TU2283
pilC'/slr0163 (Protein) - TU2283
pilJ/sll1294 (mRNA / CDS) - TU679
pilJ/sll1294 (Protein) - TU679
pilM/slr1274 (mRNA / CDS) - TU1945
pilM/slr1274 (Protein) - TU1945
pilT1/slr0161 (mRNA / CDS) - TU2283
pilT1/slr0161 (Protein) - TU2283
slr0161-5UTR (5'UTR) - TU2283
TU2283 (slr0161, slr0162, slr0163) - Transcr Unit
pilT2/sll1533 (mRNA / CDS) - TU2125
pilT2/sll1533 (Protein) - TU2125
pixG, pisG, taxP1, rer1/sll0038 (mRNA / CDS) - TU3314
pixG, pisG, taxP1, rer1/sll0038 (Protein) - TU3314
pixH, pisH/sll0039 (mRNA / CDS) - TU3311
pixH, pisH/sll0039 (Protein) - TU3311
sll0039-5UTR (5'UTR) - TU3311
TU3311 (sll0039, sll0040, sll0041, sll0042, sll0043, sll0044) - Transcr Unit
pixI, pisI/sll0040 (mRNA / CDS) - TU3311
pixI, pisI/sll0040 (Protein) - TU3311
pixJ1, pisJ1, taxD1/sll0041 (mRNA / CDS) - TU3311
pixJ1, pisJ1, taxD1/sll0041 (Protein) - TU3311
pixJ2, pisJ2/sll0042 (mRNA / CDS) - TU3311
pixJ2, pisJ2/sll0042 (Protein) - TU3311
pixL, taxAY1, hik18/sll0043 (mRNA / CDS) - TU3311
pixL, taxAY1, hik18/sll0043 (Protein) - TU3311
pknA/slr1225 (mRNA / CDS) - TU1051
pknA/slr1225 (Protein) - TU1051
pknA/slr1443 (mRNA / CDS) - TU1108
pknA/slr1443 (Protein) - TU1108
pknA/slr1697 (mRNA / CDS) - TU2066
pknA/slr1697 (Protein) - TU2066
slr1697-3UTR (3'UTR) - TU2066
slr1697-as (asRNA)
pleD/slr1047 (mRNA / CDS) - TU484
pleD/slr1047 (Protein) - TU484
slr1047-5UTR (5'UTR) - TU484
plpA, hik3/sll1124 (mRNA / CDS) - TU866
plpA, hik3/sll1124 (Protein) - TU866
sll1124-3UTR (3'UTR) - TU867
TU866 (sll1124) - Transcr Unit
plsX/slr1510 (mRNA / CDS) - TU1635
plsX/slr1510 (Protein) - TU1635
TU1635 (slr1510) - Transcr Unit
plsY/sll1973 (mRNA / CDS) - TU1610
plsY/sll1973 (Protein) - TU1610
pma1/sll1614 (mRNA / CDS) - TU1492
pma1/sll1614 (Protein) - TU1492
pmbA/slr1435 (mRNA / CDS) - TU1089
pmbA/slr1435 (Protein) - TU1089
TU914 (sll1968, sll1969) - Transcr Unit
pmgR1/ncr0700 (ncRNA) - TU1715
pnp/sll1043 (mRNA / CDS) - TU90
pnp/sll1043 (Protein) - TU90
sll1043-3UTR (3'UTR) - TU90
pntA/slr1239 (mRNA / CDS) - TU1089
pntA/slr1239 (Protein) - TU1089
slr1239-5UTR (5'UTR) - TU1089
TU1089 (slr1239, slr1240, slr1241, ssr2066, ssr2067, slr1434, slr1435) - Transcr Unit
pntB/slr1434 (mRNA / CDS) - TU1089
pntB/slr1434 (Protein) - TU1089
pobA/sll1297 (mRNA / CDS) - TU679
pobA/sll1297 (Protein) - TU679
polA/slr0707 (mRNA / CDS) - TU3271
polA/slr0707 (Protein) - TU3271
ponA/sll0002 (mRNA / CDS) - TU2609
ponA/sll0002 (Protein) - TU2609
sll0002-3UTR (3'UTR) - TU2609
sll0002-5UTR (5'UTR) - TU2609
por/slr0506 (mRNA / CDS) - TU3074
por/slr0506 (Protein) - TU3074
TU3074 (slr0506) - Transcr Unit
slr0506-3UTR (3'UTR) - TU3074
ppa/slr1622 (mRNA / CDS) - TU2111
ppa/slr1622 (Protein) - TU2111
TU2111 (slr1622) - Transcr Unit
slr1622-3UTR (3'UTR) - TU2111
ppc/sll0920 (mRNA / CDS) - TU2068
ppc/sll0920 (Protein) - TU2068
TU2068 (sll0920) - Transcr Unit
sll0920-5UTR (5'UTR) - TU2068
pphA/sll1771 (mRNA / CDS) - TU1249
pphA/sll1771 (Protein) - TU1249
ppiB/sll0227 (mRNA / CDS) - TU136
ppiB/sll0227 (Protein) - TU136
ppk/sll0290 (mRNA / CDS) - TU3169
ppk/sll0290 (Protein) - TU3169
TU3169 (sll0290) - Transcr Unit
ppnK/sll1415 (mRNA / CDS) - TU1643
ppnK/sll1415 (Protein) - TU1643
ppnK/slr0400 (mRNA / CDS) - TU2228
ppnK/slr0400 (Protein) - TU2228
TU1892 (sll1387) - Transcr Unit
ppsA/slr0301 (mRNA / CDS) - TU3180
ppsA/slr0301 (Protein) - TU3180
slr0301-3UTR (3'UTR) - TU3180
slr0301-5UTR (5'UTR) - TU3180
ppx/sll1546 (mRNA / CDS) - TU3643
ppx/sll1546 (Protein) - TU3643
TU3643 (sll1546) - Transcr Unit
sll1546-as (asRNA)
pqqE/sll0915 (mRNA / CDS) - TU2076
pqqE/sll0915 (Protein) - TU2076
prc/slr1751 (mRNA / CDS) - TU1035
prc/slr1751 (Protein) - TU1035
slr1751-5UTR (5'UTR) - TU1035
prfA/sll1110 (mRNA / CDS)
prfA/sll1110 (Protein)
prfB/sll1865 (mRNA / CDS)
prfB/sll1865 (Protein)
prfC/slr1228 (mRNA / CDS) - TU1060
prfC/slr1228 (Protein) - TU1060
prk/sll1525 (mRNA / CDS) - TU2140
prk/sll1525 (Protein) - TU2140
TU2140 (sll1525) - Transcr Unit
sll1525-3UTR (3'UTR) - TU2140
sll1525-5UTR (5'UTR) - TU2140
prlC/slr0659 (mRNA / CDS) - TU3520
prlC/slr0659 (Protein) - TU3520
prmA/sll1909 (mRNA / CDS) - TU1473
prmA/sll1909 (Protein) - TU1473
proA/sll0373 (mRNA / CDS) - TU2863
proA/sll0373 (Protein) - TU2863
proA/sll0461 (mRNA / CDS) - TU2744
proA/sll0461 (Protein) - TU2744
sll0461-5UTR (5'UTR) - TU2744
sll0461-as (asRNA)
TU2744 (sll0461, ssl0900, sll0462) - Transcr Unit
proB/slr2035 (mRNA / CDS) - TU549
proB/slr2035 (Protein) - TU549
proC/slr0661 (mRNA / CDS) - TU3521
proC/slr0661 (Protein) - TU3521
TU3521 (slr0661) - Transcr Unit
proS/sll1425 (mRNA / CDS) - TU1626
proS/sll1425 (Protein) - TU1626
TU1626 (sll1425) - Transcr Unit
prsA/sll0469 (mRNA / CDS) - TU3102
prsA/sll0469 (Protein) - TU3102
sll0469-3UTR (3'UTR) - TU3102
sll0469-5UTR (5'UTR) - TU3102
psaA/slr1834 (mRNA / CDS) - TU958
psaA/slr1834 (Protein) - TU958
TU957 (slr1834) - Transcr Unit
TU958 (slr1834) - Transcr Unit
slr1834-3UTR (3'UTR) - TU958
slr1834-5UTR (5'UTR) - TU958
psaB/slr1835 (mRNA / CDS) - TU959
psaB/slr1835 (Protein) - TU959
TU959 (slr1835) - Transcr Unit
slr1835-3UTR (3'UTR) - TU959
slr1835-5UTR (5'UTR) - TU959
psaC/ssl0563 (mRNA / CDS) - TU2375
psaC/ssl0563 (Protein) - TU2375
TU2375 (ssl0563) - Transcr Unit
ssl0563-3UTR (3'UTR) - TU2375
ssl0563-5UTR (5'UTR) - TU2375
psaD/slr0737 (mRNA / CDS) - TU113
psaD/slr0737 (Protein) - TU113
slr0737-5UTR (5'UTR) - TU113
TU113 (slr0737, slr0738) - Transcr Unit
psaE/ssr2831 (mRNA / CDS) - TU2056
psaE/ssr2831 (Protein) - TU2056
ssr2831-5UTR (5'UTR) - TU2056
TU2056 (ssr2831, slr1689, slr1690) - Transcr Unit
psaF/sll0819 (mRNA / CDS) - TU1727
psaF/sll0819 (Protein) - TU1727
sll0819-5UTR (5'UTR) - TU1727
TU1727 (sll0819, sml0008) - Transcr Unit
psaJ/sml0008 (mRNA / CDS) - TU1727
psaJ/sml0008 (Protein) - TU1727
psaK1/ssr0390 (mRNA / CDS) - TU144
psaK1/ssr0390 (Protein) - TU144
TU144 (ssr0390) - Transcr Unit
ssr0390-3UTR (3'UTR) - TU144
ssr0390-5UTR (5'UTR) - TU144
psaK2/sll0629 (mRNA / CDS) - TU3503
psaK2/sll0629 (Protein) - TU3503
psaL/slr1655 (mRNA / CDS) - TU3633
psaL/slr1655 (Protein) - TU3633
slr1655-5UTR (5'UTR) - TU3633
TU3633 (slr1655, smr0004) - Transcr Unit
TU455 (smr0005) - Transcr Unit
psb27, psbZ/slr1645 (mRNA / CDS) - TU2148
psb27, psbZ/slr1645 (Protein) - TU2148
slr1645-5UTR (5'UTR) - TU2148
TU2148 (slr1645, slr1646, slr1647) - Transcr Unit
psb13/sll1398 (mRNA / CDS) - TU46
psb28, psbW, psb13, ycf79/sll1398 (Protein) - TU46
TU46 (sll1398, sll1399, ssl2733, sll1400, sll1401) - Transcr Unit
psb28-2/slr1739 (mRNA / CDS) - TU1362
psb28-2/slr1739 (Protein) - TU1362
TU1362 (slr1739) - Transcr Unit
slr1739-3UTR (3'UTR) - TU1362
slr1739-5UTR (5'UTR) - TU1362
psbA3/sll1867 (mRNA / CDS) - TU1884
sll1867-3UTR (3'UTR) - TU1884
sll1867-5UTR (5'UTR) - TU1884
psbA1/slr1181 (mRNA / CDS) - TU3683
psbA2/slr1311 (mRNA / CDS) - TU13
psbA/slr1311,sll1867,slr1181 (Protein)
slr1311-3UTR (3'UTR) - TU13
slr1311-5UTR (5'UTR) - TU13
TU3683 (slr1181) - Transcr Unit
TU3684 (slr1181) - Transcr Unit
TU13 (slr1311) - Transcr Unit
TU1884 (sll1867) - Transcr Unit
psbB/slr0906 (mRNA / CDS) - TU2928
psbB/slr0906 (Protein) - TU2928
TU2928 (slr0906) - Transcr Unit
slr0906-3UTR (3'UTR) - TU2928
slr0906-5UTR (5'UTR) - TU2928
psbC/sll0851 (mRNA / CDS) - TU1386
psbC/sll0851 (Protein) - TU1386
psbD/sll0849 (mRNA / CDS) - TU1386
sll0849-5UTR (5'UTR) - TU1386
psbD2/slr0927 (mRNA / CDS) - TU3402
psbD/slr0927,sll0849 (Protein)
TU1386 (sll0849, sll0851) - Transcr Unit
psbE/ssr3451 (mRNA / CDS) - TU548
psbE/ssr3451 (Protein) - TU548
ssr3451-5UTR (5'UTR) - TU548
TU548 (ssr3451, smr0006, smr0007, smr0008) - Transcr Unit
psbF/smr0006 (mRNA / CDS) - TU548
psbF/smr0006 (Protein) - TU548
psbH/ssl2598 (mRNA / CDS) - TU1203
psbH/ssl2598 (Protein) - TU1203
TU1203 (ssl2598) - Transcr Unit
ssl2598-3UTR (3'UTR) - TU1203
ssl2598-5UTR (5'UTR) - TU1203
TU2443 (sml0001) - Transcr Unit
TU530 (sml0005, ncl0200, sll1635) - Transcr Unit
psbL/smr0007 (mRNA / CDS) - TU548
psbL/smr0007 (Protein) - TU548
TU134 (sml0003) - Transcr Unit
TU1204 (smr0009) - Transcr Unit
psbO/sll0427 (mRNA / CDS) - TU2166
psbO/sll0427 (Protein) - TU2166
psbP2/sll1418 (mRNA / CDS) - TU1640
psbP2/sll1418 (Protein) - TU1640
TU1640 (sll1418) - Transcr Unit
sll1418-5UTR (5'UTR) - TU1640
TU2512 (smr0001) - Transcr Unit
psbU/sll1194 (mRNA / CDS) - TU291
psbU/sll1194 (Protein) - TU291
sll1194-5UTR (5'UTR) - TU291
TU291 (sll1194, ssl2380) - Transcr Unit
psbV/sll0258 (mRNA / CDS) - TU2208
psbV/sll0258 (Protein) - TU2208
TU2754 (sml0002, ncl1310) - Transcr Unit
psbY/sml0007 (mRNA / CDS) - TU1298
psbY/sml0007 (Protein) - TU1298
sml0007-5UTR (5'UTR) - TU1298
TU1298 (sml0007, ssl3829, sll2005, sll2006, sll2007) - Transcr Unit
TU3627 (psiR1, sll1552, sll0720) - Transcr Unit
psrR1/ncr0690 (ncRNA) - TU1713
TU1713 (psrR1) - Transcr Unit
pstA/sll0682 (mRNA / CDS) - TU2785
pstA/sll0682 (Protein) - TU2785
pstB/sll0684 (mRNA / CDS) - TU2785
pstB/sll0684 (Protein) - TU2785
pstS/sll0680 (mRNA / CDS) - TU2785
pstS/sll0680 (Protein) - TU2785
pstS/slr1247 (Protein) - TU1428
pta/slr2132 (mRNA / CDS)
pta/slr2132 (Protein)
slr2132-as (asRNA)
pth/slr0922 (mRNA / CDS) - TU2942
pth/slr0922 (Protein) - TU2942
TU2942 (slr0922) - Transcr Unit
slr0922-5UTR (5'UTR) - TU2942
purA/sll1823 (mRNA / CDS) - TU830
purA/sll1823 (Protein) - TU830
purB/sll0421 (mRNA / CDS) - TU2176
purB/sll0421 (Protein) - TU2176
TU2176 (sll0421, sll0422) - Transcr Unit
purC/slr1226 (mRNA / CDS) - TU1053
purC/slr1226 (Protein) - TU1053
TU1053 (slr1226) - Transcr Unit
purD/slr1159 (mRNA / CDS) - TU1993
purD/slr1159 (Protein) - TU1993
purE/sll0901 (mRNA / CDS) - TU3399
purE/sll0901 (Protein) - TU3399
TU3399 (sll0901) - Transcr Unit
purF/sll0757 (mRNA / CDS) - TU2510
purF/sll0757 (Protein) - TU2510
sll0757-5UTR (5'UTR) - TU2510
TU2510 (sll0757, sll0759) - Transcr Unit
purH/slr0597 (mRNA / CDS) - TU3701
purH/slr0597 (Protein) - TU3701
purK/sll0578 (mRNA / CDS)
purK/sll0578 (Protein)
purL/sll1056 (mRNA / CDS) - TU75
purL/sll1056 (Protein) - TU75
TU75 (sll1056) - Transcr Unit
purL/slr0520 (mRNA / CDS) - TU3097
purL/slr0520 (Protein) - TU3097
purM/slr0838 (mRNA / CDS) - TU3001
purM/slr0838 (Protein) - TU3001
TU3001 (slr0838) - Transcr Unit
purN/slr0477 (mRNA / CDS) - TU2755
purN/slr0477 (Protein) - TU2755
TU2755 (slr0477, slr0479, slr0480, slr0482) - Transcr Unit
purS/slr0519 (Protein) - TU3097
purT/slr0861 (mRNA / CDS) - TU1392
purT/slr0861 (Protein) - TU1392
TU1392 (slr0861) - Transcr Unit
purU/sll0070 (mRNA / CDS) - TU2699
purU/sll0070 (Protein) - TU2699
TU2699 (sll0070, sll0071) - Transcr Unit
putA/sll1561 (mRNA / CDS) - TU2034
putA/sll1561 (Protein) - TU2034
TU2034 (sll1561) - Transcr Unit
pxcA/slr1596 (mRNA / CDS) - TU439
pxcA/slr1596 (Protein) - TU439
TU439 (slr1596) - Transcr Unit
slr1596-as (asRNA)
pykF/sll0587 (mRNA / CDS) - TU3122
pykF/sll0587 (Protein) - TU3122
pykF/sll1275 (mRNA / CDS) - TU1131
pykF/sll1275 (Protein) - TU1131
pyrA/sll0370 (mRNA / CDS) - TU2867
pyrA/sll0370 (Protein) - TU2867
TU2867 (sll0370, sll0371) - Transcr Unit
pyrB/slr1476 (mRNA / CDS) - TU1909
pyrB/slr1476 (Protein) - TU1909
TU1909 (slr1476) - Transcr Unit
slr1476-3UTR (3'UTR) - TU1909
pyrC/sll1018 (mRNA / CDS) - TU383
pyrC/sll1018 (Protein) - TU383
TU383 (sll1018) - Transcr Unit
pyrC/slr0406 (mRNA / CDS) - TU2241
pyrC/slr0406 (Protein) - TU2241
pyrD/slr1418 (mRNA / CDS) - TU3437
pyrD/slr1418 (Protein) - TU3437
TU3437 (slr1418) - Transcr Unit
pyrE/slr0185 (mRNA / CDS) - TU2429
pyrE/slr0185 (Protein) - TU2429
pyrF/sll0838 (mRNA / CDS) - TU1405
pyrF/sll0838 (Protein) - TU1405
pyrG/sll1443 (mRNA / CDS) - TU1962
pyrG/sll1443 (Protein) - TU1962
TU1962 (sll1443, sll1444, sll1446, sll1447, sll1166) - Transcr Unit
pyrH/sll0144 (mRNA / CDS) - TU2277
pyrH/sll0144 (Protein) - TU2277
TU2277 (sll0144, sll0145) - Transcr Unit
pyrR/sll0368 (mRNA / CDS) - TU2871
pyrR/sll0368 (Protein) - TU2871
pys, crtB/slr1255 (mRNA / CDS) - TU1438
pys, crtB/slr1255 (Protein) - TU1438
queA/sll0467 (mRNA / CDS)
queA/sll0467 (Protein)
queD/slr0078 (mRNA / CDS) - TU3038
queD/slr0078 (Protein) - TU3038
slr0078-5UTR (5'UTR) - TU3038
queE/sll2011 (Protein) - TU1291
queF/slr0711 (mRNA / CDS) - TU3283
queF/slr0711 (Protein) - TU3283
TU2185 (slr0448) - Transcr Unit
rapQ/sll0829 (mRNA / CDS) - TU3002
rapQ/sll0829 (Protein) - TU3002
sll0829-as (asRNA)
rbcL/slr0009 (mRNA / CDS) - TU2582
rbcL/slr0009 (Protein) - TU2582
TU2582 (slr0009) - Transcr Unit
slr0009-3UTR (3'UTR) - TU2582
slr0009-5UTR (5'UTR) - TU2582
rbcS/slr0012 (mRNA / CDS) - TU2584
rbcS/slr0012 (Protein) - TU2584
rbcX/slr0011 (mRNA / CDS) - TU2584
rbcX/slr0011 (Protein) - TU2584
slr0011-5UTR (5'UTR) - TU2584
TU2584 (slr0011, slr0012) - Transcr Unit
rbfA/sll0754 (mRNA / CDS) - TU2514
rbfA/sll0754 (Protein) - TU2514
TU3343 (sll0517) - Transcr Unit
rbp2/ssr1480 (mRNA / CDS) - TU1170
rbp2/ssr1480 (Protein) - TU1170
TU1170 (ssr1480) - Transcr Unit
ssr1480-3UTR (3'UTR) - TU1170
ssr1480-5UTR (5'UTR) - TU1170
rbp3/slr0193 (mRNA / CDS) - TU2878
rbp3/slr0193 (Protein) - TU2878
TU2878 (slr0193) - Transcr Unit
slr0193-5UTR (5'UTR) - TU2878
rbp1,rbpA/sll0517 (mRNA / CDS) - TU3343
rbpA/sll0517 (Protein) - TU3343
sll0517-3UTR (3'UTR) - TU3343
sll0517-5UTR (5'UTR) - TU3343
rcp1/slr0474 (mRNA / CDS) - TU2746
rcp1/slr0474 (Protein) - TU2746
slr0474-as2 (asRNA)
recA/sll0569 (mRNA / CDS) - TU3698
recA/sll0569 (Protein) - TU3698
TU3698 (sll0569, ncl1780) - Transcr Unit
TU2607 (slr0020) - Transcr Unit
recJ/sll1354 (mRNA / CDS) - TU1113
recJ/sll1354 (Protein) - TU1113
TU1113 (sll1354) - Transcr Unit
recN/sll1520 (mRNA / CDS)
recN/sll1520 (Protein)
TU991 (slr1536) - Transcr Unit
recR/slr1426 (mRNA / CDS) - TU3454
recR/slr1426 (Protein) - TU3454
TU3454 (slr1426) - Transcr Unit
rfbA/sll0207 (mRNA / CDS) - TU2634
rfbA/sll0207 (Protein) - TU2634
rfbA/sll0574 (mRNA / CDS)
rfbA/sll0574 (Protein)
rfbB/sll0575 (mRNA / CDS)
rfbB/sll0575 (Protein)
rfbB/slr0809 (mRNA / CDS) - TU1734
rfbB/slr0809 (Protein) - TU1734
rfbB/slr0836 (mRNA / CDS) - TU2994
rfbB/slr0836 (Protein) - TU2994
rfbB/slr0982 (mRNA / CDS) - TU353
rfbB/slr0982 (Protein) - TU353
TU1734 (slr0809, slr0810) - Transcr Unit
rfbC/slr0985 (mRNA / CDS) - TU353
rfbC/slr0985 (Protein) - TU353
rfbC/slr1933 (Protein)
rfbD/sll1212 (mRNA / CDS) - TU9
rfbD/sll1212 (Protein) - TU9
rfbD/sll1395 (mRNA / CDS) - TU50
rfbD/sll1395 (Protein) - TU50
rfbE/slr1615 (mRNA / CDS) - TU355
rfbE/slr1615 (Protein) - TU355
rfbF/slr0983 (mRNA / CDS) - TU353
rfbF/slr0983 (Protein) - TU353
rfbG/slr0984 (mRNA / CDS) - TU353
rfbG/slr0984 (Protein) - TU353
rfbJ/sll1457 (mRNA / CDS) - TU1016
rfbJ/sll1457 (Protein) - TU1016
rfbM/sll1370 (mRNA / CDS) - TU1926
rfbM/sll1370 (Protein) - TU1925
TU1925 (sll1370) - Transcr Unit
rfbM/slr0493 (mRNA / CDS) - TU2771
rfbM/slr0493 (Protein) - TU2771
slr0493-3UTR (3'UTR) - TU2771
rfbU/slr1064 (mRNA / CDS) - TU355
rfbU/slr1064 (Protein) - TU355
rfbW/sll0380 (mRNA / CDS) - TU2847
rfbW/sll0380 (Protein) - TU2847
rffM/slr1271 (mRNA / CDS) - TU1942
rffM/slr1271 (Protein) - TU1942
ribA/sll1894 (mRNA / CDS) - TU1829
ribA/sll1894 (Protein) - TU1829
ribC/sll0300 (mRNA / CDS) - TU2369
ribC/sll0300 (Protein) - TU2369
TU2369 (sll0300, sll0301) - Transcr Unit
ribG/slr0066 (mRNA / CDS) - TU2730
ribD/slr0066 (Protein) - TU2730
ribF/slr1882 (mRNA / CDS) - TU1269
ribF/slr1882 (Protein) - TU1269
ribH/sll1282 (mRNA / CDS) - TU1125
ribH/sll1282 (Protein) - TU1125
rimM/slr0808 (mRNA / CDS) - TU1732
rimM/slr0808 (Protein) - TU1732
rimO/slr0082 (mRNA / CDS) - TU3045
rimO/slr0082 (Protein) - TU3045
slr0082-5UTR (5'UTR) - TU3045
rimP/slr0742 (mRNA / CDS) - TU119
rimP/slr0742 (Protein) - TU119
slr0742-5UTR (5'UTR) - TU119
rlmN/sll0098 (mRNA / CDS) - TU3151
rlmN/sll0098 (Protein) - TU3151
rnb/sll1290 (mRNA / CDS) - TU683
rnb/sll1290 (Protein) - TU683
rnc/slr0346 (mRNA / CDS) - TU2530
rnc/slr0346 (Protein) - TU2530
rnc/slr1646 (mRNA / CDS) - TU2148
rnc/slr1646 (Protein) - TU2148
rnd/sll0320 (mRNA / CDS) - TU2535
rnd/sll0320 (Protein) - TU2535
rne/slr1129 (mRNA / CDS) - TU87
rne/slr1129 (Protein) - TU87
rnhA/slr0080 (mRNA / CDS) - TU3041
rnhA/slr0080 (Protein) - TU3041
TU3041 (slr0080) - Transcr Unit
rnj/slr0551 (mRNA / CDS) - TU3419
rnj/slr0551 (Protein) - TU3419
rnpA/slr1469 (mRNA / CDS) - TU1898
rnpA/slr1469 (Protein) - TU1898
rnpB/6803s01 (ncRNA) - TU139
TU139 (rnpB, slr0249) - Transcr Unit
rpaA, ycf27/slr0115 (mRNA / CDS) - TU3162
rpaA, ycf27/slr0115 (Protein) - TU3162
TU3162 (slr0115, slr0116) - Transcr Unit
rpaB, ycf27/slr0947 (mRNA / CDS) - TU2084
rpaB, ycf27/slr0947 (Protein) - TU2084
TU2084 (slr0947, slr0948) - Transcr Unit
rpe/sll0807 (mRNA / CDS) - TU1763
rpe/sll0807 (Protein) - TU1763
sll0807-5UTR (5'UTR) - TU1763
TU1763 (sll0807, SyR48) - Transcr Unit
rpiA/slr0194 (mRNA / CDS) - TU2879
rpiA/slr0194 (Protein) - TU2879
TU2879 (slr0194) - Transcr Unit
rpl1/sll1744 (mRNA / CDS) - TU931
rpl1/sll1744 (Protein) - TU931
sll1744-as (asRNA)
sll1744-as2 (asRNA)
rpl10/sll1745 (mRNA / CDS) - TU931
rpl10/sll1745 (Protein) - TU931
rpl11/sll1743 (mRNA / CDS) - TU931
rpl11/sll1743 (Protein) - TU931
rpl12/sll1746 (mRNA / CDS) - TU931
rpl12/sll1746 (Protein) - TU931
rpl13/sll1821 (mRNA / CDS) - TU833
rpl13/sll1821 (Protein) - TU833
rpl14/sll1806 (mRNA / CDS) - TU837
rpl14/sll1806 (Protein) - TU837
rpl15/sll1813 (mRNA / CDS) - TU837
rpl15/sll1813 (Protein) - TU837
rpl16/sll1805 (mRNA / CDS) - TU837
rpl16/sll1805 (Protein) - TU837
rpl17/sll1819 (mRNA / CDS) - TU833
rpl17/sll1819 (Protein) - TU833
rpl18/sll1811 (mRNA / CDS) - TU837
rpl18/sll1811 (Protein) - TU837
rpl19/sll1740 (mRNA / CDS) - TU934
rpl19/sll1740 (Protein) - TU934
TU934 (sll1740, 6803t09) - Transcr Unit
rpl2/sll1802 (mRNA / CDS) - TU837
rpl2/sll1802 (Protein) - TU837
rpl20/sll0767 (mRNA / CDS) - TU2501
rpl20/sll0767 (Protein) - TU2501
rpl21/slr1678 (mRNA / CDS) - TU2027
rpl21/slr1678 (Protein) - TU2027
rpl22/sll1803 (mRNA / CDS) - TU837
rpl22/sll1803 (Protein) - TU837
rpl23/sll1801 (mRNA / CDS) - TU837
rpl23/sll1801 (Protein) - TU837
rpl24/sll1807 (mRNA / CDS) - TU837
rpl24/sll1807 (Protein) - TU837
rpl25/sll1824 (mRNA / CDS) - TU830
rpl25/sll1824 (Protein) - TU830
rpl27/ssr2799 (mRNA / CDS) - TU2028
rpl27/ssr2799 (Protein) - TU2028
ssr2799-5UTR (5'UTR) - TU2028
TU2028 (ssr2799, slr1679) - Transcr Unit
rpl28/ssr1604 (mRNA / CDS) - TU314
rpl28/ssr1604 (Protein) - TU314
TU314 (ssr1604) - Transcr Unit
ssr1604-3UTR (3'UTR) - TU314
ssr1604-5UTR (5'UTR) - TU314
rpl29/ssl3436 (mRNA / CDS) - TU757
rpl29/ssl3436 (Protein) - TU757
rpl3/sll1799 (mRNA / CDS) - TU837
rpl3/sll1799 (Protein) - TU837
rpl31/ssl3445 (mRNA / CDS) - TU833
rpl31/ssl3445 (Protein) - TU833
rpl32/ssr1736 (mRNA / CDS) - TU488
rpl32/ssr1736 (Protein) - TU488
TU488 (ssr1736, slr1050) - Transcr Unit
rpl33/ssr1398 (mRNA / CDS) - TU2985
rpl33/ssr1398 (Protein) - TU2985
TU2985 (ssr1398, ssr1399) - Transcr Unit
TU1898 (smr0011, slr1469, slr1470, slr1471, slr1472) - Transcr Unit
rpl35/ssl1426 (mRNA / CDS) - TU2501
rpl35/ssl1426 (Protein) - TU2501
TU2501 (ssl1426, sll0767) - Transcr Unit
rpl36/sml0006 (mRNA / CDS) - TU833
rpl36/sml0006 (Protein) - TU833
sml0006-5UTR (5'UTR) - TU833
rpl4/sll1800 (mRNA / CDS) - TU837
rpl4/sll1800 (Protein) - TU837
sll1800-as (asRNA)
rpl5/sll1808 (mRNA / CDS) - TU837
rpl5/sll1808 (Protein) - TU837
rpl6/sll1810 (mRNA / CDS) - TU837
rpl6/sll1810 (Protein) - TU837
rpl9/sll1244 (mRNA / CDS) - TU1808
rpl9/sll1244 (Protein) - TU1808
TU1808 (sll1244) - Transcr Unit
sll1244-3UTR (3'UTR) - TU1808
rpoA/sll1818 (mRNA / CDS) - TU833
rpoA/sll1818 (Protein) - TU833
rpoB/sll1787 (mRNA / CDS) - TU865
rpoB/sll1787 (Protein) - TU865
rpoC1/slr1265 (mRNA / CDS) - TU1933
rpoC1/slr1265 (Protein) - TU1933
TU1933 (slr1265) - Transcr Unit
slr1265-3UTR (3'UTR) - TU1933
rpoC2/sll1789 (mRNA / CDS) - TU865
rpoC2/sll1789 (Protein) - TU865
rps10/sll1101 (mRNA / CDS) - TU1986
rps10/sll1101 (Protein) - TU1986
rps11/sll1817 (mRNA / CDS) - TU833
rps11/sll1817 (Protein) - TU833
rps12/sll1096 (mRNA / CDS) - TU1986
rps12/sll1096 (Protein) - TU1986
TU1986 (sll1096, sll1097, sll1098, sll1099, sll1101) - Transcr Unit
rps13/sll1816 (mRNA / CDS) - TU833
rps13/sll1816 (Protein) - TU833
rps14/slr0628 (mRNA / CDS) - TU2793
rps14/slr0628 (Protein) - TU2793
TU2793 (slr0628) - Transcr Unit
slr0628-3UTR (3'UTR) - TU2793
rps15/ssl1784 (mRNA / CDS) - TU332
rps15/ssl1784 (Protein) - TU332
TU332 (ssl1784, sll0933, ncl0110) - Transcr Unit
rps16/ssr0482 (mRNA / CDS) - TU2205
rps16/ssr0482 (Protein) - TU2205
ssr0482-5UTR (5'UTR) - TU2205
TU2205 (ssr0482, slr0287, ncr1020) - Transcr Unit
rps17/ssl3437 (mRNA / CDS) - TU837
rps17/ssl3437 (Protein) - TU837
rps18/ssr1399 (mRNA / CDS) - TU2985
rps18/ssr1399 (Protein) - TU2985
rps19/ssl3432 (mRNA / CDS) - TU837
rps19/ssl3432 (Protein) - TU837
rps1a/slr1356 (mRNA / CDS) - TU1129
rps1a/slr1356 (Protein) - TU1129
slr1356-5UTR (5'UTR) - TU1129
TU1129 (slr1356, ncr0450, slr1357) - Transcr Unit
rps2/sll1260 (mRNA / CDS) - TU1778
rps2/sll1260 (Protein) - TU1778
TU1778 (sll1260) - Transcr Unit
sll1260-3UTR (3'UTR) - TU1778
sll1260-5UTR (5'UTR) - TU1778
rps20/ssl2233 (mRNA / CDS) - TU865
rps20/ssl2233 (Protein) - TU865
ssl2233-5UTR (5'UTR) - TU865
TU865 (ssl2233, sll1786, sll1787, sll1789) - Transcr Unit
rps21/ssl0601 (mRNA / CDS) - TU2550
rps21/ssl0601 (Protein) - TU2550
TU2550 (ssl0601) - Transcr Unit
ssl0601-3UTR (3'UTR) - TU2550
ssl0601-5UTR (5'UTR) - TU2550
rps3/sll1804 (mRNA / CDS) - TU837
rps3/sll1804 (Protein) - TU837
rps4/slr0469 (mRNA / CDS) - TU2743
rps4/slr0469 (Protein) - TU2743
TU2743 (slr0469) - Transcr Unit
slr0469-3UTR (3'UTR) - TU2743
slr0469-5UTR (5'UTR) - TU2743
rps5/sll1812 (mRNA / CDS) - TU837
rps5/sll1812 (Protein) - TU837
rps6/sll1767 (mRNA / CDS) - TU1258
rps6/sll1767 (Protein) - TU1258
TU1258 (sll1767) - Transcr Unit
sll1767-3UTR (3'UTR) - TU1258
sll1767-5UTR (5'UTR) - TU1258
rps7/sll1097 (mRNA / CDS) - TU1986
rps7/sll1097 (Protein) - TU1986
rps8/sll1809 (mRNA / CDS) - TU837
rps8/sll1809 (Protein) - TU837
rps9/sll1822 (mRNA / CDS) - TU833
rps9/sll1822 (Protein) - TU833
rsbU/slr2031 (mRNA / CDS) - TU786
rsbU/slr2031 (Protein) - TU786
slr2031-as2 (asRNA)
rsfS/slr1886 (mRNA / CDS) - TU1277
rsfS/slr1886 (Protein) - TU1277
slr1886-3UTR (3'UTR) - TU1277
slr1886-5UTR (5'UTR) - TU1277
rsmE/slr0722 (mRNA / CDS) - TU102
rsmE/slr0722 (Protein) - TU102
rubA/slr2033 (mRNA / CDS) - TU545
rubA/slr2033 (Protein) - TU545
slr2033-5UTR (5'UTR) - TU545
TU545 (slr2033, slr2034) - Transcr Unit
ruvA/sll0876 (mRNA / CDS)
ruvA/sll0876 (Protein)
TU2803 (sll0613, sll0614) - Transcr Unit
sac1/sll0640 (mRNA / CDS) - TU413
sac1/sll0640 (Protein) - TU413
samp/slr0593 (mRNA / CDS)
samp/slr0593 (Protein)
sat/slr1165 (mRNA / CDS) - TU2006
sat/slr1165 (Protein) - TU2006
sbpA/slr1452 (mRNA / CDS)
sbpA/slr1452 (Protein)
sbtA/slr1512 (mRNA / CDS) - TU1642
sbtA/slr1512 (Protein) - TU1642
slr1512-5UTR (5'UTR) - TU1642
TU1642 (slr1512, slr1513) - Transcr Unit
SDHA/slr1233 (mRNA / CDS) - TU1067
SDHA/slr1233 (Protein) - TU1067
slr1233-5UTR (5'UTR) - TU1067
sdhB/sll0823 (mRNA / CDS) - TU3017
sdhB/sll0823 (Protein) - TU3017
sdhB/sll1625 (mRNA / CDS) - TU1353
sdhB/sll1625 (Protein) - TU1353
sds/slr0611 (Protein) - TU3720
TU3720 (slr0611) - Transcr Unit
slr0611-1 (mRNA / CDS) - TU3720
slr0611-2 (mRNA / CDS) - TU3720
secA/sll0616 (mRNA / CDS) - TU2797
secA/sll0616 (Protein) - TU2797
TU2797 (sll0616) - Transcr Unit
sll0616-3UTR (3'UTR) - TU2797
secD/slr0774 (mRNA / CDS) - TU2496
secD/slr0774 (Protein) - TU2496
TU2496 (slr0774, slr0775) - Transcr Unit
secE/ssl3335 (mRNA / CDS) - TU931
secE/ssl3335 (Protein) - TU931
ssl3335-5UTR (5'UTR) - TU931
secF/slr0775 (mRNA / CDS) - TU2496
secF/slr0775 (Protein) - TU2496
TU2341 (ssr3307) - Transcr Unit
secY/sll1814 (mRNA / CDS) - TU836
secY/sll1814 (Protein) - TU836
sll1814-5UTR (5'UTR) - TU836
TU836 (sll1814, sll1815) - Transcr Unit
serA/sll1908 (mRNA / CDS) - TU1473
serA/sll1908 (Protein) - TU1473
sll1908-5UTR (5'UTR) - TU1473
TU1473 (sll1908, sll1909) - Transcr Unit
serS/slr1703 (mRNA / CDS) - TU712
serS/slr1703 (Protein) - TU712
slr1703-as (asRNA)
sfsA/sll2014 (Protein) - TU1288
shc/slr2089 (mRNA / CDS) - TU1585
shc/slr2089 (Protein) - TU1585
sigA,rpoDI/slr0653 (mRNA / CDS) - TU3504
sigA,rpoDI/slr0653 (Protein) - TU3504
sigB/sll0306 (mRNA / CDS) - TU2364
TU2364 (sll0306) - Transcr Unit
sigB/sll0306,REV__sll0722 (Protein)
sll0306-3UTR (3'UTR) - TU2364
sll0306-5UTR (5'UTR) - TU2364
sigC/sll0184 (mRNA / CDS) - TU2887
sigC/sll0184 (Protein) - TU2887
TU2887 (sll0184) - Transcr Unit
sigE,rpoD2-V/sll1689 (mRNA / CDS) - TU1338
sigE,rpoD2-V/sll1689 (Protein) - TU1338
sigF/slr1564 (mRNA / CDS) - TU3552
sigF/slr1564 (Protein) - TU3552
TU3552 (slr1564) - Transcr Unit
slr1564-5UTR (5'UTR) - TU3552
sigG/slr1545 (mRNA / CDS) - TU1002
sigG/slr1545 (Protein) - TU1002
slr1545-5UTR (5'UTR) - TU1002
TU1001 (slr1545, slr1546, slr1547) - Transcr Unit
TU1002 (slr1545, slr1546, slr1547) - Transcr Unit
TU1380 (sll0856, sll0857, sll0858) - Transcr Unit
TU2783 (sll0687) - Transcr Unit
sir/slr0963 (mRNA / CDS) - TU320
sir/slr0963 (Protein) - TU320
TU320 (slr0963, slr0964) - Transcr Unit
SKI2/slr0451 (mRNA / CDS) - TU3671
SKI2/slr0451 (Protein) - TU3671
TU2609 (sll0002) - Transcr Unit
TU2606 (sll0005) - Transcr Unit
TU2604 (sll0006) - Transcr Unit
TU2603 (sll0007, sll0008) - Transcr Unit
TU2589 (sll0012) - Transcr Unit
TU2576 (sll0016) - Transcr Unit
TU2571 (sll0019) - Transcr Unit
TU2570 (sll0020) - Transcr Unit
TU2569 (sll0021) - Transcr Unit
TU2567 (sll0022, sll0023, sll0024) - Transcr Unit
TU3336 (sll0027) - Transcr Unit
TU3328 (sll0031) - Transcr Unit
TU3324 (sll0034) - Transcr Unit
TU3321 (sll0036) - Transcr Unit
TU3322 (sll0036) - Transcr Unit
TU3320 (sll0037) - Transcr Unit
TU3308 (sll0045) - Transcr Unit
TU3301 (sll0048, ncl1580) - Transcr Unit
TU2736 (sll0051) - Transcr Unit
TU2731 (sll0055) - Transcr Unit
TU2733 (sll0055) - Transcr Unit
TU2716 (sll0062, sll0063) - Transcr Unit
TU2714 (sll0063a, sll0064) - Transcr Unit
TU2704 (sll0068) - Transcr Unit
TU2703 (sll0069) - Transcr Unit
TU3036 (sll0083) - Transcr Unit
TU3031 (sll0086) - Transcr Unit
TU3027 (sll0088) - Transcr Unit
TU3163 (sll0092, ssl0172) - Transcr Unit
TU3153 (sll0096) - Transcr Unit
TU3151 (sll0098, sll0099) - Transcr Unit
TU3150 (sll0100, sll0101) - Transcr Unit
TU3148 (sll0102) - Transcr Unit
TU3147 (sll0103) - Transcr Unit
TU2286 (sll0136) - Transcr Unit
TU2282 (sll0141) - Transcr Unit
TU2281 (sll0142) - Transcr Unit
TU2274 (sll0147) - Transcr Unit
TU2272 (sll0148) - Transcr Unit
TU2271 (sll0149) - Transcr Unit
TU2431 (sll0154) - Transcr Unit
TU2427 (sll0156) - Transcr Unit
TU2425 (sll0157) - Transcr Unit
TU2408 (sll0167, sll0168, ncl1150) - Transcr Unit
TU2402 (sll0171) - Transcr Unit
TU2398 (sll0172, sll0173, sll0174, sll0175) - Transcr Unit
TU2396 (sll0176, sll0177) - Transcr Unit
TU2914 (sll0178) - Transcr Unit
TU2896 (sll0180, ssl0350, sll0181, sll0182) - Transcr Unit
TU2893 (sll0183) - Transcr Unit
TU2874 (sll0188, sll0189) - Transcr Unit
TU2658 (sll0191, sll0192) - Transcr Unit
TU2654 (sll0195) - Transcr Unit
TU2644 (sll0200, sll0201) - Transcr Unit
TU2643 (sll0202) - Transcr Unit
TU2639 (sll0204) - Transcr Unit
TU2623 (sll0209) - Transcr Unit
TU2617 (sll0210) - Transcr Unit
TU164 (sll0216) - Transcr Unit
TU156 (sll0217, sll0218, sll0219) - Transcr Unit
TU148 (sll0222) - Transcr Unit
TU137 (sll0224, sll0225) - Transcr Unit
TU128 (sll0230) - Transcr Unit
TU1569 (sll0236, ssl0438, sll0237, sll0238, sll0240, sll0241) - Transcr Unit
TU1567 (sll0242, sll0243) - Transcr Unit
TU1561 (sll0244) - Transcr Unit
TU1559 (sll0245) - Transcr Unit
TU1543 (sll0252) - Transcr Unit
TU1540 (sll0253) - Transcr Unit
TU1538 (sll0254) - Transcr Unit
TU2204 (sll0260) - Transcr Unit
TU2202 (sll0261) - Transcr Unit
TU2195 (sll0263) - Transcr Unit
TU2194 (sll0264, sll0265, sll0266) - Transcr Unit
TU2188 (sll0271) - Transcr Unit
TU2187 (sll0272, sll0273) - Transcr Unit
TU2186 (sll0274) - Transcr Unit
TU3192 (sll0284) - Transcr Unit
TU3193 (sll0284) - Transcr Unit
TU2388 (sll0293, sll0294) - Transcr Unit
TU2371 (sll0297, sll0298) - Transcr Unit
TU2557 (sll0312) - Transcr Unit
TU2548 (sll0314) - Transcr Unit
TU2539 (sll0318) - Transcr Unit
TU2535 (sll0319, sll0320) - Transcr Unit
TU2532 (sll0321, sll0322) - Transcr Unit
TU2524 (sll0325) - Transcr Unit
TU2481 (sll0327, sll0328, ncl1200) - Transcr Unit
TU2480 (sll0329, ncl1190) - Transcr Unit
TU2441 (sll0350) - Transcr Unit
TU2238 (sll0354) - Transcr Unit
TU2237 (sll0355) - Transcr Unit
TU2225 (sll0359) - Transcr Unit
TU2223 (sll0360) - Transcr Unit
TU2871 (sll0368, sll0369) - Transcr Unit
TU2863 (sll0372, sll0373, sll0374) - Transcr Unit
TU2858 (sll0375) - Transcr Unit
TU2855 (sll0376) - Transcr Unit
TU2854 (sll0377) - Transcr Unit
TU2847 (sll0379, sll0380) - Transcr Unit
TU2838 (sll0381, sll0382, sll0383, sll0384, sll0385) - Transcr Unit
TU2832 (sll0394, sll0395) - Transcr Unit
TU2831 (sll0396, sll0397, sll0398) - Transcr Unit
TU2692 (sll0400, sll0401) - Transcr Unit
TU2676 (sll0405, sll0406) - Transcr Unit
TU2672 (sll0408, sll0409) - Transcr Unit
TU2670 (sll0410, sll0412) - Transcr Unit
TU2669 (sll0413, ssl0787, ssl0788) - Transcr Unit
TU2665 (sll0414, sll0415) - Transcr Unit
TU2181 (sll0418) - Transcr Unit
TU2179 (sll0419, sll0420) - Transcr Unit
TU2174 (sll0423, sll0424) - Transcr Unit
TU2168 (sll0426, 6803t23) - Transcr Unit
TU2772 (sll0456) - Transcr Unit
TU3102 (sll0469) - Transcr Unit
TU3094 (sll0473) - Transcr Unit
TU3083 (sll0477, sll0478, sll0479) - Transcr Unit
TU3077 (sll0480) - Transcr Unit
TU3075 (sll0481, sll0482) - Transcr Unit
TU3068 (sll0484, sll0485, sll0486) - Transcr Unit
TU3067 (sll0487) - Transcr Unit
TU3065 (sll0488) - Transcr Unit
TU3393 (sll0496) - Transcr Unit
TU3391 (sll0497) - Transcr Unit
TU3389 (sll0498) - Transcr Unit
TU3377 (sll0505, sll0506) - Transcr Unit
TU3374 (sll0507) - Transcr Unit
TU3354 (sll0513) - Transcr Unit
TU3349 (sll0514) - Transcr Unit
TU3340 (sll0518) - Transcr Unit
TU3342 (sll0518) - Transcr Unit
TU3429 (sll0524, ssl1004) - Transcr Unit
TU3427 (sll0525) - Transcr Unit
TU3422 (sll0528) - Transcr Unit
TU3421 (sll0529) - Transcr Unit
TU3416 (sll0533) - Transcr Unit
TU3412 (sll0536, sll0537) - Transcr Unit
TU2980 (sll0539) - Transcr Unit
TU2968 (sll0542) - Transcr Unit
TU2958 (sll0546, sll0547) - Transcr Unit
TU2955 (sll0549) - Transcr Unit
TU2954 (sll0550) - Transcr Unit
TU2950 (sll0553) - Transcr Unit
TU3702 (sll0565) - Transcr Unit
TU3129 (sll0585) - Transcr Unit
TU3123 (sll0587) - Transcr Unit
TU3121 (sll0588) - Transcr Unit
TU3115 (sll0590) - Transcr Unit
TU3108 (sll0593) - Transcr Unit
TU3107 (sll0594, sll0595) - Transcr Unit
TU3106 (sll0596) - Transcr Unit
TU3105 (sll0597) - Transcr Unit
TU2826 (sll0601) - Transcr Unit
TU2819 (sll0602, sll0603) - Transcr Unit
TU2807 (sll0611) - Transcr Unit
TU2800 (sll0615) - Transcr Unit
TU3527 (sll0623) - Transcr Unit
TU3501 (sll0630) - Transcr Unit
TU426 (sll0638) - Transcr Unit
TU427 (sll0638) - Transcr Unit
TU424 (sll0639) - Transcr Unit
TU415 (sll0640) - Transcr Unit
TU406 (sll0645, sll0646) - Transcr Unit
TU405 (sll0647) - Transcr Unit
TU403 (sll0648, sll0649) - Transcr Unit
TU402 (sll0650, sll0651) - Transcr Unit
TU388 (sll0654, ncl0120, sll0656) - Transcr Unit
TU3292 (sll0657) - Transcr Unit
TU3277 (sll0667) - Transcr Unit
TU3258 (sll0677) - Transcr Unit
TU2789 (sll0679) - Transcr Unit
TU2785 (sll0680, sll0681, sll0682, sll0683, sll0684) - Transcr Unit
TU2784 (sll0685, sll0686) - Transcr Unit
TU2782 (sll0688) - Transcr Unit
TU2777 (sll0691) - Transcr Unit
TU100 (sll0708) - Transcr Unit
TU97 (sll0709, sll0710, sll0711) - Transcr Unit
TU93 (sll0716) - Transcr Unit
TU3621 (sll0721) - Transcr Unit
TU3622 (sll0721) - Transcr Unit
TU3620 (sll0722) - Transcr Unit
TU3617 (sll0723) - Transcr Unit
TU3613 (sll0723) - Transcr Unit
TU3610 (sll0726) - Transcr Unit
TU3595 (sll0733) - Transcr Unit
TU3590 (sll0735, sll0736) - Transcr Unit
TU3299 (sll0740) - Transcr Unit
TU3296 (sll0741, sll0742, sll0743, sll0744, sll0745) - Transcr Unit
TU2521 (sll0749) - Transcr Unit
TU2503 (sll0765, sll0766) - Transcr Unit
TU2493 (sll0772) - Transcr Unit
TU3253 (sll0775, sll0776, sll0777) - Transcr Unit
TU3248 (sll0779) - Transcr Unit
TU3250 (sll0779) - Transcr Unit
TU3247 (sll0781) - Transcr Unit
TU3241 (sll0783, sll0784) - Transcr Unit
TU3230 (sll0788, sll0789, sll0790) - Transcr Unit
TU3228 (sll0790) - Transcr Unit
TU1773 (sll0802, sll0803, sll0804) - Transcr Unit
TU1750 (sll0810, sll0811) - Transcr Unit
TU1740 (sll0814, ncl0820) - Transcr Unit
TU3019 (sll0821) - Transcr Unit
TU3017 (sll0822, sll0823) - Transcr Unit
TU3010 (sll0825) - Transcr Unit
TU3005 (sll0827, sll0828) - Transcr Unit
TU3002 (sll0829) - Transcr Unit
TU2991 (sll0833) - Transcr Unit
TU2989 (sll0834) - Transcr Unit
TU1405 (sll0837, sll0838, sll0839) - Transcr Unit
TU1402 (sll0842) - Transcr Unit
TU1398 (sll0843) - Transcr Unit
TU1397 (sll0844) - Transcr Unit
TU1394 (sll0846) - Transcr Unit
TU1390 (sll0847) - Transcr Unit
TU1381 (sll0853, sll0854, sll0855) - Transcr Unit
TU1193 (sll0867, sll0868) - Transcr Unit
TU1180 (sll0872) - Transcr Unit
TU1161 (sll0877) - Transcr Unit
TU2917 (sll0886) - Transcr Unit
TU2916 (sll0887) - Transcr Unit
TU3405 (sll0899, sll0900) - Transcr Unit
TU2103 (sll0905) - Transcr Unit
TU2090 (sll0909, sll0910, sll0911) - Transcr Unit
TU2087 (sll0912) - Transcr Unit
TU2085 (sll0913) - Transcr Unit
TU2076 (sll0915) - Transcr Unit
TU2067 (sll0921) - Transcr Unit
TU344 (sll0923) - Transcr Unit
TU343 (sll0924) - Transcr Unit
TU338 (sll0925) - Transcr Unit
TU337 (sll0926) - Transcr Unit
TU335 (sll0927) - Transcr Unit
TU327 (sll0936, ssl1792) - Transcr Unit
TU316 (sll0943) - Transcr Unit
TU2361 (sll0944) - Transcr Unit
TU665 (sll0980, sll0981) - Transcr Unit
TU664 (sll0982, sll0983) - Transcr Unit
TU662 (sll0984) - Transcr Unit
TU633 (sll0993, sll0994, sll0995) - Transcr Unit
TU631 (sll0996) - Transcr Unit
TU630 (sll0997) - Transcr Unit
TU500 (sll1004) - Transcr Unit
TU498 (sll1006) - Transcr Unit
TU487 (sll1011) - Transcr Unit
TU378 (sll1020) - Transcr Unit
TU377 (sll1021) - Transcr Unit
TU374 (sll1022, sll1023, sll1024) - Transcr Unit
TU198 (sll1035, ssl2009, sll1036, sll1037) - Transcr Unit
TU196 (sll1037) - Transcr Unit
TU184 (sll1039) - Transcr Unit
TU178 (sll1040, sll1041, sll1318) - Transcr Unit
TU90 (sll1043) - Transcr Unit
TU83 (sll1049) - Transcr Unit
TU78 (sll1053, sll1054) - Transcr Unit
TU67 (sll1061, sll1062) - Transcr Unit
TU63 (sll1063) - Transcr Unit
TU819 (sll1070) - Transcr Unit
TU817 (sll1071, sll1072) - Transcr Unit
TU818 (sll1071, sll1072) - Transcr Unit
TU794 (sll1080, sll1081) - Transcr Unit
TU790 (sll1084) - Transcr Unit
TU2013 (sll1086) - Transcr Unit
TU2012 (sll1087, sll1089) - Transcr Unit
TU885 (sll1118) - Transcr Unit
TU879 (sll1119) - Transcr Unit
TU877 (sll1120) - Transcr Unit
TU876 (sll1121) - Transcr Unit
TU867 (sll1123, sll1124) - Transcr Unit
TU1081 (sll1129) - Transcr Unit
TU1074 (sll1131, sll1132) - Transcr Unit
TU1062 (sll1135) - Transcr Unit
TU1052 (sll1138) - Transcr Unit
TU1047 (sll1142) - Transcr Unit
TU1036 (sll1144) - Transcr Unit
TU1441 (sll1147) - Transcr Unit
TU1436 (sll1150, ncl0610) - Transcr Unit
TU1427 (sll1154) - Transcr Unit
TU1426 (sll1155, sll1156, sll1157) - Transcr Unit
TU1420 (sll1159, sll1160, sll1161, sll1162, sll1163, sll1164) - Transcr Unit
TU1415 (sll1165) - Transcr Unit
TU1934 (sll1178) - Transcr Unit
TU1929 (sll1180) - Transcr Unit
TU1927 (sll1181) - Transcr Unit
TU311 (sll1186) - Transcr Unit
TU310 (sll1187, sll1188) - Transcr Unit
TU297 (sll1191, sll1192) - Transcr Unit
TU290 (sll1196) - Transcr Unit
TU284 (sll1200) - Transcr Unit
TU277 (sll1201) - Transcr Unit
TU279 (sll1201) - Transcr Unit
TU24 (sll1206) - Transcr Unit
TU9 (sll1212, sll1213) - Transcr Unit
TU1720 (sll1217, sll1218) - Transcr Unit
TU1718 (sll1219) - Transcr Unit
TU1701 (sll1233) - Transcr Unit
TU1699 (sll1234) - Transcr Unit
TU1811 (sll1242) - Transcr Unit
TU1797 (sll1251) - Transcr Unit
TU1794 (sll1252, sll1253) - Transcr Unit
TU1791 (sll1253) - Transcr Unit
TU1792 (sll1253) - Transcr Unit
TU1788 (sll1254, sll1255) - Transcr Unit
TU1785 (sll1256, sll1257) - Transcr Unit
TU1776 (sll1262, sll1263) - Transcr Unit
TU1157 (sll1265) - Transcr Unit
TU1159 (sll1265) - Transcr Unit
TU1155 (sll1267) - Transcr Unit
TU1153 (sll1268) - Transcr Unit
TU1140 (sll1273) - Transcr Unit
TU1135 (sll1274) - Transcr Unit
TU1131 (sll1275, sll1276, sll1277) - Transcr Unit
TU1125 (sll1280, 6803t12, sll1281, sll1282, sll1348) - Transcr Unit
TU702 (sll1283, sll1284, sll1285) - Transcr Unit
TU696 (sll1286) - Transcr Unit
TU683 (sll1289, sll1290) - Transcr Unit
TU681 (sll1291) - Transcr Unit
TU680 (sll1292, sll1293) - Transcr Unit
TU679 (sll1294, sll1296, ssl2559, sll1297) - Transcr Unit
TU673 (sll1298, sll1299, sll1300) - Transcr Unit
TU1220 (sll1308) - Transcr Unit
TU166 (sll1321, sll1322, ssl2615, sll1323, sll1324, sll1325, sll1326, sll1327) - Transcr Unit
TU3470 (sll1329) - Transcr Unit
TU3469 (sll1330) - Transcr Unit
TU3466 (sll1333) - Transcr Unit
TU3461 (sll1334) - Transcr Unit
TU3457 (sll1336) - Transcr Unit
TU3455 (sll1338) - Transcr Unit
TU3450 (sll1341) - Transcr Unit
TU3442 (sll1343) - Transcr Unit
TU3440 (sll1344) - Transcr Unit
TU1124 (sll1349) - Transcr Unit
TU1122 (sll1350) - Transcr Unit
TU1117 (sll1352) - Transcr Unit
TU1105 (sll1355) - Transcr Unit
TU1101 (sll1356) - Transcr Unit
TU1100 (sll1358, sll1359) - Transcr Unit
TU3487 (sll1365) - Transcr Unit
TU3484 (sll1366) - Transcr Unit
TU3473 (sll1367) - Transcr Unit
TU1921 (sll1372, sll1373) - Transcr Unit
TU1915 (sll1376) - Transcr Unit
TU1911 (sll1377, sll1378) - Transcr Unit
TU1906 (sll1380, sll1381) - Transcr Unit
TU1901 (sll1384) - Transcr Unit
TU1889 (sll1388, sll1389) - Transcr Unit
TU1886 (sll1390, sll1866) - Transcr Unit
TU50 (sll1394, sll1395, sll1396, sll1397) - Transcr Unit
TU32 (sll1404, sll1405, sll1406, sll1407, sll1408) - Transcr Unit
TU1643 (sll1414, sll1415) - Transcr Unit
TU1629 (sll1424) - Transcr Unit
TU1624 (sll1426) - Transcr Unit
TU1623 (sll1427) - Transcr Unit
TU1615 (sll1429) - Transcr Unit
TU1613 (sll1430) - Transcr Unit
TU1973 (sll1434) - Transcr Unit
TU1972 (sll1435) - Transcr Unit
TU1971 (sll1436, sll1437) - Transcr Unit
TU1016 (sll1457) - Transcr Unit
TU1013 (sll1459) - Transcr Unit
TU1010 (sll1461, sll1462) - Transcr Unit
TU3574 (sll1479) - Transcr Unit
TU3572 (sll1481, sll1482) - Transcr Unit
TU3571 (sll1483, sll1484) - Transcr Unit
TU3570 (sll1485, sll1486) - Transcr Unit
TU3559 (sll1488, sll1489) - Transcr Unit
TU3557 (sll1491) - Transcr Unit
TU3546 (sll1495) - Transcr Unit
TU3544 (sll1495) - Transcr Unit
TU3543 (sll1496) - Transcr Unit
TU3538 (sll1498) - Transcr Unit
TU457 (sll1507) - Transcr Unit
TU447 (sll1512) - Transcr Unit
TU438 (sll1516) - Transcr Unit
TU2154 (sll1522) - Transcr Unit
TU2137 (sll1526) - Transcr Unit
TU2134 (sll1528, sll1530, sll1531) - Transcr Unit
TU2123 (sll1535) - Transcr Unit
TU2107 (sll1541) - Transcr Unit
TU3653 (sll1544) - Transcr Unit
TU3647 (sll1545) - Transcr Unit
TU3642 (sll1547) - Transcr Unit
TU3630 (sll1550) - Transcr Unit
TU2050 (sll1556) - Transcr Unit
TU2049 (sll1557) - Transcr Unit
TU2047 (sll1558) - Transcr Unit
TU2040 (sll1559, sll1560) - Transcr Unit
TU2033 (sll1562, sll1563) - Transcr Unit
TU2029 (sll1564) - Transcr Unit
TU749 (sll1568) - Transcr Unit
TU742 (sll1571, sll1572, ncl0300) - Transcr Unit
TU730 (sll1573, sll1574, sll1575) - Transcr Unit
TU707 (sll1581, sll1582) - Transcr Unit
TU706 (sll1583) - Transcr Unit
TU704 (sll1584) - Transcr Unit
TU1529 (sll1586) - Transcr Unit
TU1531 (sll1586) - Transcr Unit
TU1532 (sll1586) - Transcr Unit
TU1534 (sll1586) - Transcr Unit
TU1537 (sll1586) - Transcr Unit
TU1503 (sll1605, sll1606) - Transcr Unit
TU1496 (sll1611, sll1612) - Transcr Unit
TU1364 (sll1618, 6803t13) - Transcr Unit
TU1361 (sll1620) - Transcr Unit
TU1359 (sll1621) - Transcr Unit
TU1356 (sll1623) - Transcr Unit
TU1355 (sll1624, ncl0580) - Transcr Unit
TU1353 (sll1625) - Transcr Unit
TU1352 (sll1626) - Transcr Unit
TU1347 (sll1628, sll1629, sll1630, sll1631) - Transcr Unit
TU1033 (sll1632) - Transcr Unit
TU1026 (sll1634) - Transcr Unit
TU524 (sll1638) - Transcr Unit
TU519 (sll1642) - Transcr Unit
TU515 (sll1643) - Transcr Unit
TU509 (sll1647) - Transcr Unit
TU273 (Sll1651, sll1652) - Transcr Unit
TU270 (sll1654, sll1655) - Transcr Unit
TU268 (sll1656) - Transcr Unit
TU261 (sll1659) - Transcr Unit
TU259 (sll1660, ssl3222) - Transcr Unit
TU255 (sll1662) - Transcr Unit
TU253 (sll1663, sll1664) - Transcr Unit
TU251 (sll1665) - Transcr Unit
TU248 (sll1666) - Transcr Unit
TU247 (sll1667) - Transcr Unit
TU233 (sll1675) - Transcr Unit
TU231 (sll1676) - Transcr Unit
TU230 (sll1677, sll1678) - Transcr Unit
TU223 (sll1682) - Transcr Unit
TU1335 (sll1692) - Transcr Unit
TU1332 (sll1693) - Transcr Unit
TU1324 (sll1697) - Transcr Unit
TU1312 (sll1708) - Transcr Unit
TU986 (sll1712, sll1713) - Transcr Unit
TU987 (sll1712, sll1713) - Transcr Unit
TU981 (sll1716) - Transcr Unit
TU978 (sll1717) - Transcr Unit
TU966 (sll1721) - Transcr Unit
TU962 (sll1722) - Transcr Unit
TU954 (sll1730) - Transcr Unit
TU949 (sll1735) - Transcr Unit
TU948 (sll1736) - Transcr Unit
TU537 (sll1749a, sll1749, sll1750, sll1751) - Transcr Unit
TU531 (sll1752) - Transcr Unit
TU1271 (sll1757) - Transcr Unit
TU1267 (sll1758) - Transcr Unit
TU1264 (sll1762) - Transcr Unit
TU1263 (sll1763) - Transcr Unit
TU1262 (sll1764, sll1765, sll1766) - Transcr Unit
TU1253 (sll1768) - Transcr Unit
TU1248 (sll1772) - Transcr Unit
TU1246 (sll1773, ssl3389, ncl0520) - Transcr Unit
TU1239 (sll1774) - Transcr Unit
TU1241 (sll1774) - Transcr Unit
TU1237 (sll1775, sll1776) - Transcr Unit
TU1236 (sll1776) - Transcr Unit
TU1231 (sll1780) - Transcr Unit
TU1224 (sll1783, sll1784, ssl3410, sll1785, sll1304, sll1305, sll1306, sll1307) - Transcr Unit
TU863 (sll1791) - Transcr Unit
TU828 (sll1825) - Transcr Unit
TU621 (sll1830) - Transcr Unit
TU618 (sll1831) - Transcr Unit
TU616 (sll1832, sll1833) - Transcr Unit
TU608 (sll1834) - Transcr Unit
TU606 (sll1835) - Transcr Unit
TU602 (sll1837) - Transcr Unit
TU2343 (sll1841) - Transcr Unit
TU2332 (sll1845) - Transcr Unit
TU2322 (sll1848, sll1849) - Transcr Unit
TU2319 (sll1852, sll1853) - Transcr Unit
TU2315 (sll1858) - Transcr Unit
TU2309 (sll1861) - Transcr Unit
TU2304 (sll1862) - Transcr Unit
TU2303 (sll1863) - Transcr Unit
TU2295 (sll1865) - Transcr Unit
TU1876 (sll1869, sll1870) - Transcr Unit
TU1873 (sll1872) - Transcr Unit
TU1872 (sll1873) - Transcr Unit
TU1867 (sll1878) - Transcr Unit
TU1865 (sll1880) - Transcr Unit
TU1862 (sll1882) - Transcr Unit
TU1859 (sll1884) - Transcr Unit
TU1854 (sll1885) - Transcr Unit
TU1853 (sll1886) - Transcr Unit
TU1835 (sll1890) - Transcr Unit
TU1837 (sll1890) - Transcr Unit
TU1834 (sll1891) - Transcr Unit
TU1832 (sll1892, ncl0870, sll1893) - Transcr Unit
TU1821 (sll1897) - Transcr Unit
TU1818 (sll1898, sll1899) - Transcr Unit
TU1479 (sll1906) - Transcr Unit
TU783 (sll1911) - Transcr Unit
TU774 (sll1915) - Transcr Unit
TU755 (sll1925) - Transcr Unit
TU752 (sll1926) - Transcr Unit
TU750 (sll1927) - Transcr Unit
TU593 (sll1930) - Transcr Unit
TU586 (sll1934) - Transcr Unit
TU569 (sll1937) - Transcr Unit
TU568 (sll1938) - Transcr Unit
TU564 (sll1939, sll1940) - Transcr Unit
TU556 (sll1942) - Transcr Unit
TU542 (sll1945) - Transcr Unit
TU1469 (sll1949) - Transcr Unit
TU1467 (sll1950) - Transcr Unit
TU1463 (sll1951) - Transcr Unit
TU1465 (sll1951) - Transcr Unit
TU1466 (sll1951) - Transcr Unit
TU1459 (sll1954) - Transcr Unit
TU1450 (sll1960) - Transcr Unit
TU1449 (sll1961) - Transcr Unit
TU909 (sll1971) - Transcr Unit
TU1610 (sll1973) - Transcr Unit
TU1598 (sll1979) - Transcr Unit
TU1590 (sll1982) - Transcr Unit
TU1589 (sll1983, sll1984, sll1985) - Transcr Unit
TU1669 (sll1997, sll1998, sll1999) - Transcr Unit
TU1660 (sll2001, ssl2749, sll1411) - Transcr Unit
TU1303 (sll2002) - Transcr Unit
TU1307 (sll2002) - Transcr Unit
TU1295 (sll2008, sll2009) - Transcr Unit
TU1286 (sll2015) - Transcr Unit
TU5001 (sll5002, ssl5001) - Transcr Unit
TU5003 (sll5004, sll5003) - Transcr Unit
TU5021 (sll5006) - Transcr Unit
TU5031 (sll5028, ssl5027) - Transcr Unit
TU5036 (sll5036, sll5035, sll5034, sll5033, sll5032, ssl5031) - Transcr Unit
TU5046 (sll5041) - Transcr Unit
TU5051 (sll5046, ssl5045, sll5044, sll5043) - Transcr Unit
TU5054 (sll5048) - Transcr Unit
TU5056 (sll5049) - Transcr Unit
TU5058 (sll5052) - Transcr Unit
TU5064 (sll5057) - Transcr Unit
TU5069 (sll5059) - Transcr Unit
TU5073 (sll5061, sll5060) - Transcr Unit
TU5077 (sll5063) - Transcr Unit
TU5082 (sll5069) - Transcr Unit
TU5089 (sll5076) - Transcr Unit
TU5092 (sll5080, sll5079) - Transcr Unit
TU5096 (sll5083) - Transcr Unit
TU5129 (sll5128) - Transcr Unit
TU6005 (sll6010) - Transcr Unit
TU6016 (sll6017) - Transcr Unit
TU6049 (sll6055, sll6054, sll6053) - Transcr Unit
TU6060 (sll6069) - Transcr Unit
TU6071 (sll6076) - Transcr Unit
TU6086 (sll6093, ssl6092) - Transcr Unit
TU6092 (sll6098) - Transcr Unit
TU6101 (sll6109) - Transcr Unit
TU7015 (sll7027) - Transcr Unit
TU7016 (sll7028) - Transcr Unit
TU7026 (sll7034, sll7033) - Transcr Unit
TU7035 (sll7044, sll7043, ssl7042) - Transcr Unit
TU7043 (sll7047) - Transcr Unit
TU7046 (sll7050) - Transcr Unit
TU7052 (sll7056) - Transcr Unit
TU7058 (sll7067, sll7066, sll7065, sll7064, sll7063, sll7062) - Transcr Unit
TU7065 (sll7070, sll7069) - Transcr Unit
TU7071 (sll7075, ssl7074) - Transcr Unit
TU7078 (sll7087, sll7086, sll7085) - Transcr Unit
TU7079 (sll7090, sll7089) - Transcr Unit
TU8002 (sll8001) - Transcr Unit
TU8005 (sll8007, sll8006, ssl8005, sll8004, ssl8003, sll8002) - Transcr Unit
TU8008 (sll8012, sll8011, ssl8010) - Transcr Unit
TU8012 (sll8017) - Transcr Unit
TU8014 (sll8018) - Transcr Unit
TU8016 (sll8018) - Transcr Unit
TU8019 (sll8019) - Transcr Unit
TU8041 (sll8043, sll8042, ssl8041) - Transcr Unit
TU8043 (sll8048) - Transcr Unit
TU2568 (slr0001) - Transcr Unit
TU2578 (slr0006, slr0007) - Transcr Unit
TU2587 (slr0013) - Transcr Unit
TU2588 (slr0014) - Transcr Unit
TU2592 (slr0015) - Transcr Unit
TU2595 (slr0016, ncr1270) - Transcr Unit
TU2601 (slr0019) - Transcr Unit
TU2608 (slr0021) - Transcr Unit
TU3306 (slr0022) - Transcr Unit
TU3310 (slr0023) - Transcr Unit
TU3316 (slr0031) - Transcr Unit
TU3317 (slr0032) - Transcr Unit
TU3318 (slr0033) - Transcr Unit
TU2696 (slr0049, ssr0102, slr0050) - Transcr Unit
TU2712 (slr0054, slr0055) - Transcr Unit
TU2718 (slr0059, slr0060, slr0061) - Transcr Unit
TU2729 (slr0065) - Transcr Unit
TU2734 (slr0067) - Transcr Unit
TU3038 (slr0078) - Transcr Unit
TU3043 (slr0081, ncr1500, ncr1510) - Transcr Unit
TU3045 (slr0082) - Transcr Unit
TU3053 (slr0086) - Transcr Unit
TU3057 (slr0090) - Transcr Unit
TU3060 (slr0096, slr0498) - Transcr Unit
TU3145 (slr0104) - Transcr Unit
TU3152 (slr0108) - Transcr Unit
TU3155 (slr0110, slr0111) - Transcr Unit
TU3159 (slr0112) - Transcr Unit
TU3160 (slr0114) - Transcr Unit
TU3168 (slr0121) - Transcr Unit
TU2264 (slr0144, slr0145, slr0146, slr0147, slr0148, slr0149, slr0150, slr0151) - Transcr Unit
TU2267 (slr0152) - Transcr Unit
TU2270 (slr0152) - Transcr Unit
TU2393 (slr0169) - Transcr Unit
TU2395 (slr0172) - Transcr Unit
TU2414 (slr0179, slr0180) - Transcr Unit
TU2419 (slr0181) - Transcr Unit
TU2429 (slr0185) - Transcr Unit
TU2876 (slr0191, slr0192) - Transcr Unit
TU2880 (slr0195) - Transcr Unit
TU2883 (slr0197) - Transcr Unit
TU2891 (slr0201) - Transcr Unit
TU2902 (slr0207) - Transcr Unit
TU2903 (slr0207) - Transcr Unit
TU2904 (slr0208) - Transcr Unit
TU2911 (slr0211) - Transcr Unit
TU2622 (slr0217, ncr1290) - Transcr Unit
TU2648 (slr0226) - Transcr Unit
TU2659 (slr0230) - Transcr Unit
TU2660 (slr0232) - Transcr Unit
TU122 (slr0237, slr0238) - Transcr Unit
TU126 (slr0240, slr0241) - Transcr Unit
TU130 (slr0244) - Transcr Unit
TU135 (slr0245) - Transcr Unit
TU140 (slr0250) - Transcr Unit
TU141 (slr0250) - Transcr Unit
TU1549 (slr0265) - Transcr Unit
TU1552 (slr0265) - Transcr Unit
TU1563 (slr0270) - Transcr Unit
TU1565 (slr0271, slr0272, slr0273) - Transcr Unit
TU2189 (slr0280) - Transcr Unit
TU2203 (slr0284) - Transcr Unit
TU2212 (slr0292) - Transcr Unit
TU2213 (slr0293) - Transcr Unit
TU2215 (slr0294, slr0392, slr0393) - Transcr Unit
TU3174 (slr0298, slr0299) - Transcr Unit
TU3180 (slr0301) - Transcr Unit
TU3190 (slr0306) - Transcr Unit
TU3200 (slr0312) - Transcr Unit
TU3202 (slr0316) - Transcr Unit
TU3207 (slr0319) - Transcr Unit
TU2363 (slr0320) - Transcr Unit
TU2370 (slr0326, slr0327, slr0328, slr0329) - Transcr Unit
TU2380 (slr0333, slr0334, ssr0536) - Transcr Unit
TU2387 (slr0337) - Transcr Unit
TU2391 (slr0338, slr0168) - Transcr Unit
TU2523 (slr0341) - Transcr Unit
TU2530 (slr0345, ssr0550, slr0346) - Transcr Unit
TU2534 (slr0347) - Transcr Unit
TU2538 (slr0348) - Transcr Unit
TU2540 (slr0350) - Transcr Unit
TU2543 (slr0351) - Transcr Unit
TU2545 (slr0352) - Transcr Unit
TU2561 (slr0358) - Transcr Unit
TU2436 (slr0360) - Transcr Unit
TU2437 (slr0361) - Transcr Unit
TU2449 (slr0364) - Transcr Unit
TU2451 (slr0366) - Transcr Unit
TU2453 (slr0368, slr0369) - Transcr Unit
TU2456 (slr0370) - Transcr Unit
TU2462 (slr0373, slr0374) - Transcr Unit
TU2463 (slr0376, ncr1200) - Transcr Unit
TU2464 (slr0377) - Transcr Unit
TU2466 (slr0378, slr0379) - Transcr Unit
TU2469 (slr0380) - Transcr Unit
TU2472 (slr0381) - Transcr Unit
TU2484 (slr0386) - Transcr Unit
TU2226 (slr0398) - Transcr Unit
TU2228 (slr0400, slr0401, slr0402) - Transcr Unit
TU2233 (slr0404) - Transcr Unit
TU2243 (slr0408) - Transcr Unit
TU2835 (slr0416) - Transcr Unit
TU2849 (slr0420, slr0421) - Transcr Unit
TU2859 (slr0423, ncr1390, ssr0706) - Transcr Unit
TU2668 (slr0431) - Transcr Unit
TU2683 (slr0438) - Transcr Unit
TU2684 (slr0439) - Transcr Unit
TU2695 (slr0440) - Transcr Unit
TU2158 (slr0442) - Transcr Unit
TU2169 (slr0443) - Transcr Unit
TU2178 (slr0445) - Transcr Unit
TU3671 (slr0451) - Transcr Unit
TU3673 (slr0454) - Transcr Unit
TU3674 (slr0454) - Transcr Unit
TU3675 (slr0455, slr0456) - Transcr Unit
TU2752 (slr0476) - Transcr Unit
TU2753 (slr0476) - Transcr Unit
TU2758 (slr0482) - Transcr Unit
TU2760 (slr0483) - Transcr Unit
TU2769 (slr0488) - Transcr Unit
TU2767 (slr0488) - Transcr Unit
TU2768 (slr0488) - Transcr Unit
TU2770 (slr0489, slr0491, slr0492) - Transcr Unit
TU2771 (slr0493) - Transcr Unit
TU3085 (slr0513) - Transcr Unit
TU3089 (slr0516) - Transcr Unit
TU3095 (slr0517) - Transcr Unit
TU3096 (slr0518) - Transcr Unit
TU3097 (slr0519, slr0520, slr0521) - Transcr Unit
TU3103 (slr0523) - Transcr Unit
TU3359 (slr0534) - Transcr Unit
TU3363 (slr0535, slr0536) - Transcr Unit
TU3371 (slr0537) - Transcr Unit
TU3388 (slr0544) - Transcr Unit
TU3420 (slr0554) - Transcr Unit
TU3424 (slr0556) - Transcr Unit
TU2953 (slr0565) - Transcr Unit
TU2957 (slr0569) - Transcr Unit
TU2961 (slr0572) - Transcr Unit
TU2965 (slr0574) - Transcr Unit
TU2967 (slr0575) - Transcr Unit
TU2972 (slr0580) - Transcr Unit
TU2975 (slr0580) - Transcr Unit
TU2976 (slr0581, slr0582) - Transcr Unit
TU2982 (slr0583) - Transcr Unit
TU3690 (slr0587) - Transcr Unit
TU3691 (slr0589, slr0590) - Transcr Unit
TU3694 (slr0591) - Transcr Unit
TU3699 (slr0594) - Transcr Unit
TU3701 (slr0596, slr0597) - Transcr Unit
TU3703 (slr0598, slr0599) - Transcr Unit
TU3704 (slr0599) - Transcr Unit
TU3705 (slr0599) - Transcr Unit
TU3706 (slr0600) - Transcr Unit
TU3707 (slr0601) - Transcr Unit
TU3708 (slr0602) - Transcr Unit
TU3711 (slr0604) - Transcr Unit
TU3713 (slr0605) - Transcr Unit
TU3714 (slr0606) - Transcr Unit
TU3715 (slr0607) - Transcr Unit
TU3717 (slr0609, slr0610) - Transcr Unit
TU2 (slr0612) - Transcr Unit
TU4 (slr0613) - Transcr Unit
TU3110 (slr0615, ncr1520) - Transcr Unit
TU3112 (slr0616) - Transcr Unit
TU3114 (slr0617) - Transcr Unit
TU3119 (slr0619, ssr1041) - Transcr Unit
TU3132 (slr0624) - Transcr Unit
TU3133 (slr0625) - Transcr Unit
TU3135 (slr0626, slr0099) - Transcr Unit
TU2796 (slr0630) - Transcr Unit
TU2806 (slr0634, slr0635) - Transcr Unit
TU2811 (slr0637, slr0638, slr0639, slr0640) - Transcr Unit
TU2813 (slr0642) - Transcr Unit
TU2816 (slr0643) - Transcr Unit
TU2818 (slr0644) - Transcr Unit
TU2821 (slr0645, slr0646) - Transcr Unit
TU3499 (slr0651, slr0652) - Transcr Unit
TU3512 (slr0656) - Transcr Unit
TU3516 (slr0657, slr0658, ncr1670) - Transcr Unit
TU3520 (slr0659) - Transcr Unit
TU3525 (slr0662) - Transcr Unit
TU3530 (slr0665, slr0666) - Transcr Unit
TU3532 (slr0667, slr0668) - Transcr Unit
TU398 (slr0679) - Transcr Unit
TU399 (slr0680, slr0681) - Transcr Unit
TU418 (slr0688) - Transcr Unit
TU3257 (slr0702) - Transcr Unit
TU3274 (slr0708) - Transcr Unit
TU3282 (slr0709) - Transcr Unit
TU3285 (slr0712) - Transcr Unit
TU2790 (slr0719) - Transcr Unit
TU95 (slr0721) - Transcr Unit
TU103 (slr0723) - Transcr Unit
TU104 (slr0724, slr0725, slr0727) - Transcr Unit
TU107 (slr0729, slr0730, slr0731) - Transcr Unit
TU111 (slr0734) - Transcr Unit
TU116 (slr0740) - Transcr Unit
TU118 (slr0741) - Transcr Unit
TU119 (slr0742, slr0743, slr0743a, slr0744, slr0236) - Transcr Unit
TU3596 (slr0751) - Transcr Unit
TU3597 (slr0752) - Transcr Unit
TU3608 (slr0755, ssr1258, ssr1260) - Transcr Unit
TU3295 (slr0765) - Transcr Unit
TU2489 (slr0769, slr0770, slr0771) - Transcr Unit
TU2492 (slr0773) - Transcr Unit
TU2497 (slr0776) - Transcr Unit
TU2499 (slr0776) - Transcr Unit
TU2500 (slr0776) - Transcr Unit
TU2511 (slr0779) - Transcr Unit
TU2513 (slr0780) - Transcr Unit
TU2517 (slr0782) - Transcr Unit
TU3211 (slr0787) - Transcr Unit
TU3212 (slr0789, slr0790) - Transcr Unit
TU1725 (slr0804, slr0806) - Transcr Unit
TU1728 (slr0807) - Transcr Unit
TU1733 (slr0808) - Transcr Unit
TU1745 (slr0813) - Transcr Unit
TU1754 (slr0818, slr0819) - Transcr Unit
TU1759 (slr0820) - Transcr Unit
TU1762 (slr0821) - Transcr Unit
TU1765 (slr0822) - Transcr Unit
TU1767 (slr0822) - Transcr Unit
TU1769 (slr0822) - Transcr Unit
TU2983 (slr0825) - Transcr Unit
TU2986 (slr0827, ncr1460) - Transcr Unit
TU2994 (slr0835, slr0836) - Transcr Unit
TU3006 (slr0841, ncr1470, slr0842) - Transcr Unit
TU3016 (slr0845) - Transcr Unit
TU3024 (slr0848) - Transcr Unit
TU1377 (slr0852, slr0853) - Transcr Unit
TU1384 (slr0856, slr0857) - Transcr Unit
TU1395 (slr0862) - Transcr Unit
TU1396 (slr0863) - Transcr Unit
TU1403 (slr0864) - Transcr Unit
TU1409 (slr0869, slr0870, slr0871, ssr1473, slr0872) - Transcr Unit
TU1163 (slr0875) - Transcr Unit
TU1167 (slr0876) - Transcr Unit
TU1171 (slr0877) - Transcr Unit
TU1174 (slr0879, slr0880) - Transcr Unit
TU1178 (slr0881) - Transcr Unit
TU1179 (slr0881) - Transcr Unit
TU1184 (slr0883) - Transcr Unit
TU1195 (slr0887) - Transcr Unit
TU1196 (slr0888, slr0889) - Transcr Unit
TU2921 (slr0897) - Transcr Unit
TU2929 (slr0907, slr0909, slr0912, slr0913, slr0914) - Transcr Unit
TU2937 (slr0918, slr0919, slr0920) - Transcr Unit
TU2944 (slr0924) - Transcr Unit
TU3403 (slr0929, slr0930, ssr1558) - Transcr Unit
TU2072 (slr0935, slr0936) - Transcr Unit
TU2074 (slr0937) - Transcr Unit
TU2075 (slr0938) - Transcr Unit
TU2079 (slr0942) - Transcr Unit
TU2081 (slr0944) - Transcr Unit
TU2100 (slr0957) - Transcr Unit
TU2105 (slr0959) - Transcr Unit
TU2106 (slr0960) - Transcr Unit
TU315 (slr0962) - Transcr Unit
TU325 (slr0967) - Transcr Unit
TU330 (slr0971) - Transcr Unit
TU342 (slr0975, 6803t05) - Transcr Unit
TU349 (slr0976, slr0977) - Transcr Unit
TU350 (slr0978) - Transcr Unit
TU352 (slr0980, slr0981) - Transcr Unit
TU353 (slr0982, slr0983, slr0984, slr0985, slr1610, slr1611) - Transcr Unit
TU2349 (slr0989) - Transcr Unit
TU2350 (slr0990, slr0992) - Transcr Unit
TU628 (slr1019) - Transcr Unit
TU640 (slr1024, slr1025) - Transcr Unit
TU647 (slr1028) - Transcr Unit
TU653 (slr1028) - Transcr Unit
TU663 (slr1037) - Transcr Unit
TU479 (slr1041) - Transcr Unit
TU481 (slr1042, slr1043, slr1044, slr1045, slr1046) - Transcr Unit
TU484 (slr1047, slr1048) - Transcr Unit
TU490 (slr1051, slr1052) - Transcr Unit
TU497 (slr1056) - Transcr Unit
TU362 (slr1075, slr1076) - Transcr Unit
TU364 (slr1077) - Transcr Unit
TU366 (slr1079) - Transcr Unit
TU369 (slr1081, slr1082, slr1083, slr1084, slr1085, slr1087) - Transcr Unit
TU379 (slr1090) - Transcr Unit
TU183 (slr1094) - Transcr Unit
TU185 (slr1095) - Transcr Unit
TU186 (slr1096, slr1097, slr1098, slr1099) - Transcr Unit
TU188 (slr1100, slr1101, slr1102) - Transcr Unit
TU191 (slr1103, slr1104) - Transcr Unit
TU195 (slr1105) - Transcr Unit
TU199 (slr1106) - Transcr Unit
TU200 (slr1107) - Transcr Unit
TU212 (slr1109, slr1110, slr1772) - Transcr Unit
TU57 (slr1113, slr1114, slr1115, slr1116, slr1117) - Transcr Unit
TU64 (slr1119) - Transcr Unit
TU65 (slr1119) - Transcr Unit
TU71 (slr1120) - Transcr Unit
TU77 (slr1122) - Transcr Unit
TU80 (slr1124, slr1125) - Transcr Unit
TU84 (slr1127, slr1128) - Transcr Unit
TU800 (slr1142, slr1143) - Transcr Unit
TU806 (slr1145) - Transcr Unit
TU1982 (slr1152) - Transcr Unit
TU1994 (slr1160) - Transcr Unit
TU1999 (slr1161, slr1162) - Transcr Unit
TU2000 (slr1163) - Transcr Unit
TU2003 (slr1164) - Transcr Unit
TU2006 (slr1165, slr1166) - Transcr Unit
TU2015 (slr1174, SyR49) - Transcr Unit
TU3679 (slr1176, slr1177) - Transcr Unit
TU3680 (slr1178, slr1179) - Transcr Unit
TU3685 (slr1186) - Transcr Unit
TU3686 (slr1187, slr1188, slr1189) - Transcr Unit
TU3687 (slr1192) - Transcr Unit
TU881 (slr1194, slr1195, slr1196, slr1197) - Transcr Unit
TU886 (slr1198, slr1199) - Transcr Unit
TU888 (slr1200, slr1201) - Transcr Unit
TU889 (slr1202) - Transcr Unit
TU890 (slr1203) - Transcr Unit
TU891 (slr1204) - Transcr Unit
TU893 (slr1205, slr1206, slr1207, ncr0370) - Transcr Unit
TU896 (slr1208, slr1209, slr1210) - Transcr Unit
TU907 (slr1215) - Transcr Unit
TU1048 (slr1223) - Transcr Unit
TU1049 (slr1224) - Transcr Unit
TU1051 (slr1225) - Transcr Unit
TU1055 (slr1227) - Transcr Unit
TU1060 (slr1228, slr1229, slr1230) - Transcr Unit
TU1063 (slr1232) - Transcr Unit
TU1067 (slr1233, ssr2049) - Transcr Unit
TU1069 (slr1234) - Transcr Unit
TU1070 (slr1235, slr1236) - Transcr Unit
TU1417 (slr1243) - Transcr Unit
TU1428 (slr1247, slr1248, slr1249, slr1250) - Transcr Unit
TU1430 (slr1251) - Transcr Unit
TU1437 (slr1253) - Transcr Unit
TU1445 (slr1258) - Transcr Unit
TU1446 (slr1259, slr1260, slr1261, slr1262) - Transcr Unit
TU1448 (slr1263) - Transcr Unit
TU1938 (slr1266, slr1267, slr1269) - Transcr Unit
TU1942 (slr1270, slr1271) - Transcr Unit
TU1944 (slr1272, slr1273, ncr0830, slr1274, slr1275, slr1276) - Transcr Unit
TU1945 (slr1272, slr1273, ncr0830, slr1274, slr1275, slr1276) - Transcr Unit
TU1946 (slr1277) - Transcr Unit
TU1951 (slr1278) - Transcr Unit
TU1956 (slr1282, slr1283) - Transcr Unit
TU295 (slr1299) - Transcr Unit
TU296 (slr1300) - Transcr Unit
TU302 (slr1301) - Transcr Unit
TU306 (slr1305) - Transcr Unit
TU309 (slr1306) - Transcr Unit
TU14 (slr1312) - Transcr Unit
TU17 (slr1315) - Transcr Unit
TU23 (slr1318, slr1319) - Transcr Unit
TU1690 (slr1324) - Transcr Unit
TU1696 (slr1325, ncr0680) - Transcr Unit
TU1697 (slr1325, ncr0680) - Transcr Unit
TU1717 (slr1334) - Transcr Unit
TU1781 (slr1336) - Transcr Unit
TU1786 (slr1338, slr1339, slr1340) - Transcr Unit
TU1790 (slr1342, slr1343) - Transcr Unit
TU1796 (slr1344) - Transcr Unit
TU1800 (slr1347, slr1348, SYR51) - Transcr Unit
TU1802 (slr1349) - Transcr Unit
TU1810 (slr1353) - Transcr Unit
TU1134 (slr1362) - Transcr Unit
TU1136 (slr1363) - Transcr Unit
TU1150 (slr1367) - Transcr Unit
TU671 (slr1376, slr1377) - Transcr Unit
TU672 (slr1378, slr1379, slr1380) - Transcr Unit
TU677 (slr1385, ssr2318) - Transcr Unit
TU695 (slr1394, ssr2340) - Transcr Unit
TU698 (slr1396) - Transcr Unit
TU700 (slr1397) - Transcr Unit
TU701 (slr1398) - Transcr Unit
TU1215 (slr1403) - Transcr Unit
TU1216 (slr1403) - Transcr Unit
TU1219 (slr1403) - Transcr Unit
TU1223 (slr1406, slr1407, slr1409, slr1410) - Transcr Unit
TU165 (slr1411) - Transcr Unit
TU169 (slr1413) - Transcr Unit
TU172 (slr1416) - Transcr Unit
TU3438 (slr1419, slr1420) - Transcr Unit
TU3460 (slr1428) - Transcr Unit
TU3467 (slr1431) - Transcr Unit
TU1091 (slr1437, slr1438) - Transcr Unit
TU1098 (slr1440) - Transcr Unit
TU1108 (slr1442, slr1443) - Transcr Unit
TU1109 (slr1444) - Transcr Unit
TU1118 (slr1448) - Transcr Unit
TU1120 (slr1449, slr1450) - Transcr Unit
TU3471 (slr1451) - Transcr Unit
TU3474 (slr1455) - Transcr Unit
TU3486 (slr1461) - Transcr Unit
TU3488 (slr1462) - Transcr Unit
TU3490 (slr1462) - Transcr Unit
TU3494 (slr1464) - Transcr Unit
TU1895 (slr1467) - Transcr Unit
TU1914 (slr1478) - Transcr Unit
TU37 (slr1484, slr1485, slr1488) - Transcr Unit
TU39 (slr1489, slr1490) - Transcr Unit
TU49 (slr1495, slr1496) - Transcr Unit
TU55 (slr1501) - Transcr Unit
TU56 (slr1501) - Transcr Unit
TU1621 (slr1503) - Transcr Unit
TU1622 (slr1503) - Transcr Unit
TU1632 (slr1506, ncr0640) - Transcr Unit
TU1633 (slr1507) - Transcr Unit
TU1634 (slr1508, slr1509) - Transcr Unit
TU1653 (slr1519, slr1520) - Transcr Unit
TU1654 (slr1521) - Transcr Unit
TU1655 (slr1522) - Transcr Unit
TU1659 (slr1524) - Transcr Unit
TU1969 (slr1530) - Transcr Unit
TU1980 (slr1533, slr1534) - Transcr Unit
TU989 (slr1535) - Transcr Unit
TU995 (slr1540, slr1541, slr1542) - Transcr Unit
TU997 (slr1543) - Transcr Unit
TU1014 (slr1549) - Transcr Unit
TU3535 (slr1556) - Transcr Unit
TU3537 (slr1556) - Transcr Unit
TU3540 (slr1557) - Transcr Unit
TU3554 (slr1565, slr1566) - Transcr Unit
TU3560 (slr1570, slr1571, slr1572) - Transcr Unit
TU3562 (slr1573) - Transcr Unit
TU3567 (slr1576, slr1577) - Transcr Unit
TU3568 (slr1579) - Transcr Unit
TU3578 (slr1583) - Transcr Unit
TU3581 (slr1584, slr1585) - Transcr Unit
TU3582 (slr1584, slr1585) - Transcr Unit
TU3586 (slr1588, slr0746) - Transcr Unit
TU435 (slr1593, slr1594, slr1595) - Transcr Unit
TU448 (slr1598) - Transcr Unit
TU451 (slr1599) - Transcr Unit
TU454 (slr1600) - Transcr Unit
TU456 (slr1601) - Transcr Unit
TU459 (slr1603) - Transcr Unit
TU473 (slr1608) - Transcr Unit
TU476 (slr1609) - Transcr Unit
TU477 (slr1609) - Transcr Unit
TU355 (slr1612, slr1613, slr1614, slr1615, slr1616, slr1617, slr1618, slr1619, Norf2, slr1062, slr1063, slr1064, slr1065, slr1066, slr1067, slr1068, slr1069, slr1070, slr1071, slr1072, slr1073, slr1074) - Transcr Unit
TU2112 (slr1623, slr1624) - Transcr Unit
TU2113 (slr1623, slr1624) - Transcr Unit
TU2118 (slr1626) - Transcr Unit
TU2119 (slr1627) - Transcr Unit
TU2122 (slr1628, slr1629) - Transcr Unit
TU2127 (slr1634) - Transcr Unit
TU2129 (slr1635, slr1636) - Transcr Unit
TU2133 (slr1638, slr1639) - Transcr Unit
TU2146 (slr1644) - Transcr Unit
TU2147 (slr1644) - Transcr Unit
TU3628 (slr1652) - Transcr Unit
TU3629 (slr1653) - Transcr Unit
TU3638 (slr1657, slr1658, slr1659) - Transcr Unit
TU3640 (slr1660) - Transcr Unit
TU3645 (slr1661) - Transcr Unit
TU3651 (slr1666, ssr2781) - Transcr Unit
TU3658 (slr1667) - Transcr Unit
TU3661 (slr1668, ssr2786) - Transcr Unit
TU2024 (slr1674, slr1675) - Transcr Unit
TU2026 (slr1676, slr1677) - Transcr Unit
TU2041 (slr1683) - Transcr Unit
TU2043 (slr1686) - Transcr Unit
TU2045 (slr1686) - Transcr Unit
TU2046 (slr1687) - Transcr Unit
TU2058 (slr1691) - Transcr Unit
TU2062 (slr1692) - Transcr Unit
TU2063 (slr1693, slr1694) - Transcr Unit
TU2066 (slr1697) - Transcr Unit
TU703 (slr1699) - Transcr Unit
TU712 (slr1702, slr1703) - Transcr Unit
TU725 (slr1704) - Transcr Unit
TU733 (slr1705) - Transcr Unit
TU736 (slr1706) - Transcr Unit
TU741 (slr1708, ncr0320) - Transcr Unit
TU748 (slr1710) - Transcr Unit
TU1491 (slr1712) - Transcr Unit
TU1498 (slr1715, slr1716) - Transcr Unit
TU1502 (slr1717) - Transcr Unit
TU1505 (slr1718) - Transcr Unit
TU1506 (slr1719, slr1720, slr1721) - Transcr Unit
TU1508 (slr1722) - Transcr Unit
TU1509 (slr1723) - Transcr Unit
TU1513 (slr1724) - Transcr Unit
TU1520 (slr1726) - Transcr Unit
TU1524 (slr1727) - Transcr Unit
TU1345 (slr1732) - Transcr Unit
TU1357 (slr1736, slr1737) - Transcr Unit
TU1363 (slr1740) - Transcr Unit
TU1366 (slr1742) - Transcr Unit
TU1368 (slr1744) - Transcr Unit
TU1369 (slr1744) - Transcr Unit
TU1373 (slr1746, ncr0520) - Transcr Unit
TU1374 (slr1747) - Transcr Unit
TU1029 (slr1748) - Transcr Unit
TU1035 (slr1751, slr1752) - Transcr Unit
TU505 (slr1753) - Transcr Unit
TU508 (slr1755) - Transcr Unit
TU513 (slr1760) - Transcr Unit
TU514 (slr1761) - Transcr Unit
TU516 (slr1762) - Transcr Unit
TU517 (slr1763) - Transcr Unit
TU525 (slr1768) - Transcr Unit
TU216 (slr1776) - Transcr Unit
TU217 (slr1776) - Transcr Unit
TU235 (slr1788, slr1789) - Transcr Unit
TU238 (slr1790, slr1791) - Transcr Unit
TU244 (slr1793) - Transcr Unit
TU249 (slr1796) - Transcr Unit
TU257 (slr1798) - Transcr Unit
TU264 (slr1799) - Transcr Unit
TU1314 (slr1804, slr1805) - Transcr Unit
TU1317 (slr1807) - Transcr Unit
TU1321 (slr1809) - Transcr Unit
TU1325 (slr1811, slr1812) - Transcr Unit
TU1326 (slr1813, slr1814) - Transcr Unit
TU1328 (slr1815, slr1816) - Transcr Unit
TU1334 (slr1818) - Transcr Unit
TU1336 (slr1819) - Transcr Unit
TU1344 (slr1821, slr1822) - Transcr Unit
TU936 (slr1826, slr1827) - Transcr Unit
TU965 (slr1837) - Transcr Unit
TU969 (slr1840) - Transcr Unit
TU970 (slr1841) - Transcr Unit
TU971 (slr1841) - Transcr Unit
TU534 (slr1847, slr1848) - Transcr Unit
TU539 (slr1851, ssr3129) - Transcr Unit
TU1230 (slr1862) - Transcr Unit
TU1232 (slr1863, slr1864) - Transcr Unit
TU1233 (slr1865, slr1866) - Transcr Unit
TU1242 (slr1869, ssr3159) - Transcr Unit
TU1245 (slr1870, ncr0500) - Transcr Unit
TU1254 (slr1874, slr1875) - Transcr Unit
TU1260 (slr1877, slr1878) - Transcr Unit
TU1277 (slr1886) - Transcr Unit
TU829 (slr1890) - Transcr Unit
TU848 (slr1894) - Transcr Unit
TU850 (slr1895) - Transcr Unit
TU851 (slr1895) - Transcr Unit
TU852 (slr1896) - Transcr Unit
TU854 (slr1897) - Transcr Unit
TU860 (slr1902, slr1903) - Transcr Unit
TU598 (slr1906, slr1907) - Transcr Unit
TU599 (slr1908, slr1909, slr1910) - Transcr Unit
TU601 (slr1910) - Transcr Unit
TU603 (slr1911) - Transcr Unit
TU604 (slr1912) - Transcr Unit
TU605 (slr1913, slr1914) - Transcr Unit
TU612 (slr1916) - Transcr Unit
TU613 (slr1917) - Transcr Unit
TU615 (slr1918) - Transcr Unit
TU622 (slr1919) - Transcr Unit
TU624 (slr1920) - Transcr Unit
TU2293 (slr1923, slr1924, slr1925) - Transcr Unit
TU2299 (slr1926, slr1927) - Transcr Unit
TU2316 (slr1934) - Transcr Unit
TU2317 (slr1935) - Transcr Unit
TU2323 (slr1936, slr1937) - Transcr Unit
TU2324 (slr1938) - Transcr Unit
TU2325 (slr1939) - Transcr Unit
TU2328 (slr1939) - Transcr Unit
TU2331 (slr1940, ssr3300) - Transcr Unit
TU2338 (slr1945) - Transcr Unit
TU2342 (slr1946) - Transcr Unit
TU2346 (slr1949) - Transcr Unit
TU1814 (slr1956) - Transcr Unit
TU1815 (slr1957) - Transcr Unit
TU1816 (slr1958) - Transcr Unit
TU1819 (slr1959) - Transcr Unit
TU1822 (slr1960) - Transcr Unit
TU1824 (slr1962) - Transcr Unit
TU1827 (slr1963, slr1964) - Transcr Unit
TU1828 (slr1964) - Transcr Unit
TU1831 (slr1966) - Transcr Unit
TU1848 (slr1971) - Transcr Unit
TU1857 (slr1974) - Transcr Unit
TU1868 (slr1977) - Transcr Unit
TU1869 (slr1978) - Transcr Unit
TU1875 (slr1982, slr1983) - Transcr Unit
TU1476 (slr1990, slr1991) - Transcr Unit
TU1482 (slr1999, slr2000) - Transcr Unit
TU754 (slr2005) - Transcr Unit
TU756 (slr2006, slr2007) - Transcr Unit
TU757 (slr2008, slr2009, slr2010, ssr3409, ssr3410, slr2011, slr2012, slr2013) - Transcr Unit
TU770 (slr2024) - Transcr Unit
TU771 (slr2025) - Transcr Unit
TU776 (slr2027) - Transcr Unit
TU786 (slr2030, slr2031, ssr1880, slr1133, slr1134) - Transcr Unit
TU554 (slr2036) - Transcr Unit
TU561 (slr2041) - Transcr Unit
TU570 (slr2043) - Transcr Unit
TU574 (slr2045) - Transcr Unit
TU577 (slr2046) - Transcr Unit
TU580 (slr2047, ncl0215) - Transcr Unit
TU582 (slr2048, slr2049) - Transcr Unit
TU585 (slr2052) - Transcr Unit
TU587 (slr2053) - Transcr Unit
TU1454 (slr2057, ncr0530) - Transcr Unit
TU1460 (slr2062, ncr0560) - Transcr Unit
TU911 (slr2070, slr2071) - Transcr Unit
TU920 (slr2074) - Transcr Unit
TU926 (slr2077) - Transcr Unit
TU1570 (slr2079) - Transcr Unit
TU1600 (slr2095, slr2096) - Transcr Unit
TU1606 (slr2101) - Transcr Unit
TU1611 (slr2105) - Transcr Unit
TU1668 (slr2108) - Transcr Unit
TU1670 (slr2110, slr2111) - Transcr Unit
TU1672 (slr2112, slr2113, slr2114) - Transcr Unit
TU1673 (slr2115, slr2116, slr2117, slr2118) - Transcr Unit
TU1674 (slr2119) - Transcr Unit
TU1677 (slr2122, slr2123, slr2124, slr2125, slr2126) - Transcr Unit
TU1679 (slr2128) - Transcr Unit
TU1683 (slr2131, ssr3588) - Transcr Unit
TU1297 (slr2136) - Transcr Unit
TU1300 (slr2141) - Transcr Unit
TU1306 (slr2143) - Transcr Unit
TU1308 (slr2144, slr1803) - Transcr Unit
TU5005 (slr5005) - Transcr Unit
TU5011 (slr5005) - Transcr Unit
TU5012 (slr5005) - Transcr Unit
TU5019 (slr5005) - Transcr Unit
TU5022 (slr5010) - Transcr Unit
TU5034 (slr5029) - Transcr Unit
TU5037 (slr5037, slr5038) - Transcr Unit
TU5057 (slr5051) - Transcr Unit
TU5060 (slr5053, slr5054) - Transcr Unit
TU5062 (slr5055, slr5056) - Transcr Unit
TU5068 (slr5058) - Transcr Unit
TU5086 (slr5071) - Transcr Unit
TU5087 (slr5073, ssr5074) - Transcr Unit
TU5090 (slr5077) - Transcr Unit
TU5091 (slr5078) - Transcr Unit
TU5094 (slr5082) - Transcr Unit
TU5095 (slr5082) - Transcr Unit
TU5104 (slr5101, slr5102) - Transcr Unit
TU5115 (slr5110) - Transcr Unit
TU5117 (slr5111, slr5112) - Transcr Unit
TU5121 (slr5115, slr5116, ssr5117, slr5118) - Transcr Unit
TU5122 (slr5119, ssr5120) - Transcr Unit
TU5127 (slr5126, slr5127) - Transcr Unit
TU6007 (slr6011) - Transcr Unit
TU6009 (slr6012, slr6013, slr6014, slr6015, slr6016) - Transcr Unit
TU6020 (slr6021) - Transcr Unit
TU6023 (slr6022) - Transcr Unit
TU6036 (slr6037, slr6038) - Transcr Unit
TU6037 (slr6039, slr6040, slr6041) - Transcr Unit
TU6052 (slr6058) - Transcr Unit
TU6057 (slr6063, slr6064, slr6065, slr6066, slr6067, slr6068) - Transcr Unit
TU6062 (slr6070) - Transcr Unit
TU6064 (slr6071, slr6072, slr6073, slr6074, slr6075) - Transcr Unit
TU6075 (slr6080) - Transcr Unit
TU6078 (slr6081) - Transcr Unit
TU6088 (slr6095, slr6096) - Transcr Unit
TU6090 (slr6097) - Transcr Unit
TU6096 (slr6103) - Transcr Unit
TU6098 (slr6104, slr6105) - Transcr Unit
TU6104 (slr6110) - Transcr Unit
TU7004 (slr7010, slr7011, slr7012) - Transcr Unit
TU7011 (slr7023, slr7024, slr7025, slr7026) - Transcr Unit
TU7053 (slr7057, slr7058) - Transcr Unit
TU7055 (slr7061) - Transcr Unit
TU7057 (slr7061) - Transcr Unit
TU7066 (slr7071) - Transcr Unit
TU7069 (slr7073) - Transcr Unit
TU7075 (slr7080, slr7081, slr7082) - Transcr Unit
TU7087 (slr7094, slr7095) - Transcr Unit
TU7088 (slr7096) - Transcr Unit
TU7091 (slr7100) - Transcr Unit
TU7094 (slr7101) - Transcr Unit
TU8023 (slr8026) - Transcr Unit
TU8025 (slr8029) - Transcr Unit
TU8026 (slr8030) - Transcr Unit
TU8028 (slr8030) - Transcr Unit
TU8030 (slr8036) - Transcr Unit
TU8031 (slr8036) - Transcr Unit
TU8034 (slr8038) - Transcr Unit
TU8036 (slr8038) - Transcr Unit
TU8044 (slr8045, slr8046) - Transcr Unit
slt/slr0534 (mRNA / CDS)
slt/slr0534 (Protein)
slr0534-as3 (asRNA)
TU833 (sml0006, sll1816, sll1817, sll1818, sll1819, sll1820, sll1821, sll1822, ssl3445) - Transcr Unit
sml0011 (mRNA / CDS) - TU807
sml0011 (Protein) - TU807
TU807 (sml0011) - Transcr Unit
sml0011-3UTR (3'UTR) - TU807
sml0011-5UTR (5'UTR) - TU807
sml0013 (mRNA / CDS) - TU3351
sml0013 (Protein) - TU3351
sml0013-5UTR (5'UTR) - TU3351
TU3351 (sml0013, sll0514) - Transcr Unit
smpB/slr1639 (mRNA / CDS) - TU2133
smpB/slr1639 (Protein) - TU2133
sodB/slr1516 (mRNA / CDS) - TU1649
sodB/slr1516 (Protein) - TU1649
slr1516-5UTR (5'UTR) - TU1649
TU1649 (slr1516, ssr2553) - Transcr Unit
speB1/sll0228 (mRNA / CDS) - TU136
speB1/sll0228 (Protein) - TU136
speB2/sll1077 (mRNA / CDS) - TU795
speB2/sll1077 (Protein) - TU795
sphX/sll0679 (mRNA / CDS) - TU2789
sphX/sll0679 (Protein) - TU2789
spkA/sll1575 (mRNA / CDS) - TU730
spkA/sll1575 (Protein) - TU730
spkA'/sll1574 (mRNA / CDS) - TU730
spkA'/sll1574 (Protein) - TU730
spkD/sll0776 (mRNA / CDS) - TU3253
spkD/sll0776 (Protein) - TU3253
spoT/slr1325 (mRNA / CDS) - TU1697
spoT/slr1325 (Protein) - TU1697
slr1325-5UTR (5'UTR) - TU1697
SpoU/slr1673 (mRNA / CDS) - TU2022
SpoU/slr1673 (Protein) - TU2022
spsA/sll0045 (mRNA / CDS)
spsA/sll0045 (Protein)
sqdB/slr1020 (mRNA / CDS) - TU632
sqdB/slr1020 (Protein) - TU632
TU632 (slr1020) - Transcr Unit
slr1020-3UTR (3'UTR) - TU632
slr1020-5UTR (5'UTR) - TU632
sqdX/slr0384 (mRNA / CDS) - TU2477
sqdX/slr0384 (Protein) - TU2477
TU2477 (slr0384) - Transcr Unit
srrA/slr1897 (mRNA / CDS) - TU854
srrA/slr1897 (Protein) - TU854
ssaA/6S RNA/ncr0870 (ncRNA) - TU1961
TU1961 (ssaA/6S RNA) - Transcr Unit
ssb/slr0925 (mRNA / CDS) - TU3400
ssb/slr0925 (Protein) - TU3400
TU3400 (slr0925, ssr1552, slr0926) - Transcr Unit
TU2289 (ssl0241) - Transcr Unit
TU2288 (ssl0242, sll0135) - Transcr Unit
TU2279 (ssl0259) - Transcr Unit
TU2421 (ssl0294) - Transcr Unit
TU2895 (ssl0352) - Transcr Unit
TU2892 (ssl0353) - Transcr Unit
TU2637 (ssl0385, sll0205) - Transcr Unit
TU145 (ssl0410) - Transcr Unit
TU1554 (ssl0461, sll0250) - Transcr Unit
TU1542 (ssl0467) - Transcr Unit
TU2208 (ssl0483, sll0258) - Transcr Unit
TU3206 (ssl0511) - Transcr Unit
TU2373 (ssl0564, sll0296) - Transcr Unit
TU2374 (ssl0564, sll0296) - Transcr Unit
TU2686 (ssl0769) - Transcr Unit
TU2962 (ssl1046, sll0543) - Transcr Unit
TU2959 (ssl1047, sll0544, sll0545) - Transcr Unit
TU3291 (ssl1255, sll0658, sll0659) - Transcr Unit
TU3286 (ssl1263, sll0661, sll0662) - Transcr Unit
TU2778 (ssl1300, sll0690) - Transcr Unit
TU3603 (ssl1376, ssl1377) - Transcr Unit
TU3238 (ssl1464, ncl1560) - Transcr Unit
TU1775 (ssl1498) - Transcr Unit
TU1735 (ssl1520, sll0818) - Transcr Unit
TU3014 (ssl1533) - Transcr Unit
TU2996 (ssl1552) - Transcr Unit
TU1407 (ssl1577) - Transcr Unit
TU2940 (ssl1690, sll0891, sll0892) - Transcr Unit
TU2070 (ssl1762) - Transcr Unit
TU2071 (ssl1762) - Transcr Unit
TU2358 (ssl1807) - Transcr Unit
TU504 (ssl1918, sll1001) - Transcr Unit
TU371 (ssl1972, sll1025) - Transcr Unit
TU69 (ssl2069, sll1060) - Transcr Unit
TU2001 (ssl2148) - Transcr Unit
TU1983 (ssl2162) - Transcr Unit
TU1078 (ssl2245, sll1130) - Transcr Unit
TU1071 (ssl2250) - Transcr Unit
TU1429 (ssl2296) - Transcr Unit
TU289 (ssl2384, sll1198) - Transcr Unit
TU1205 (ssl2595) - Transcr Unit
TU1106 (ssl2648) - Transcr Unit
TU1888 (ssl2717) - Transcr Unit
TU1964 (ssl2807) - Transcr Unit
TU1006 (ssl2823, sll1466) - Transcr Unit
TU3563 (ssl2874) - Transcr Unit
TU2139 (ssl2971) - Transcr Unit
TU3663 (ssl2996) - Transcr Unit
TU3657 (ssl2999) - Transcr Unit
TU2044 (ssl3044) - Transcr Unit
TU1500 (ssl3142) - Transcr Unit
TU1030 (ssl3177) - Transcr Unit
TU1310 (ssl3291, sll1709, sll1710) - Transcr Unit
TU984 (ssl3297, sll1714, sll1715) - Transcr Unit
TU931 (ssl3335, sll1742, sll1743, sll1744, sll1745, sll1746) - Transcr Unit
TU1256 (ssl3379) - Transcr Unit
TU1249 (ssl3382, ssl3383, sll1769, sll1770, sll1771, sll1772) - Transcr Unit
TU827 (ssl3451, sll1068) - Transcr Unit
TU1849 (ssl3549, sll1888) - Transcr Unit
TU1823 (ssl3573) - Transcr Unit
TU780 (ssl3615, sll1912, sll1913) - Transcr Unit
TU1681 (ssl3803, sll1994, sll1995) - Transcr Unit
TU5102 (ssl5098, sll5097, ssl5096) - Transcr Unit
TU5119 (ssl5114, ssl5113) - Transcr Unit
TU6079 (ssl6082) - Transcr Unit
TU7001 (ssl7004, sll7003) - Transcr Unit
TU7002 (ssl7007, sll7006) - Transcr Unit
TU7033 (ssl7039, ssl7038) - Transcr Unit
TU2711 (ssr0109, slr0053) - Transcr Unit
TU2910 (ssr0332) - Transcr Unit
TU2624 (ssr0349) - Transcr Unit
TU2235 (ssr0657) - Transcr Unit
TU2240 (ssr0663) - Transcr Unit
TU2842 (ssr0692) - Transcr Unit
TU2844 (ssr0693) - Transcr Unit
TU2162 (ssr0755, ssr0756) - Transcr Unit
TU2163 (ssr0757, ssr0759) - Transcr Unit
TU3113 (ssr1038) - Transcr Unit
TU2795 (ssr1049) - Transcr Unit
TU3529 (ssr1114, slr0664) - Transcr Unit
TU3254 (ssr1169) - Transcr Unit
TU3265 (ssr1175, slr0703, smr0002, slr0704, ssr1176) - Transcr Unit
TU3606 (ssr1256) - Transcr Unit
TU1723 (ssr1375) - Transcr Unit
TU2993 (ssr1407, slr0833) - Transcr Unit
TU2926 (ssr1528) - Transcr Unit
TU3408 (ssr1562) - Transcr Unit
TU365 (ssr1765, ssr1766, slr1078) - Transcr Unit
TU70 (ssr1853) - Transcr Unit
TU895 (ssr2016) - Transcr Unit
TU1065 (ssr2047) - Transcr Unit
TU1066 (ssr2047) - Transcr Unit
TU1083 (ssr2061, slr1238, ncr0430) - Transcr Unit
TU1084 (ssr2062) - Transcr Unit
TU1421 (ssr2078, slr1246) - Transcr Unit
TU1960 (ssr2130, slr1287, slr1288) - Transcr Unit
TU299 (ssr2153) - Transcr Unit
TU300 (ssr2153) - Transcr Unit
TU1693 (ssr2194) - Transcr Unit
TU1702 (ssr2201, slr1327) - Transcr Unit
TU674 (ssr2315, slr1383, ssr2317, slr1384) - Transcr Unit
TU689 (ssr2333, slr1392) - Transcr Unit
TU3439 (ssr2377, slr1421, ncr1610) - Transcr Unit
TU1094 (ssr2406) - Transcr Unit
TU1110 (ssr2422) - Transcr Unit
TU1644 (ssr2549) - Transcr Unit
TU1651 (ssr2554, slr1518) - Transcr Unit
TU1015 (ssr2611, slr1550) - Transcr Unit
TU469 (ssr2708, ssr2710, ssr2711) - Transcr Unit
TU2109 (ssr2723) - Transcr Unit
TU2153 (ssr2754, ssr2755, slr1649) - Transcr Unit
TU3656 (ssr2784) - Transcr Unit
TU3665 (ssr2787) - Transcr Unit
TU2032 (ssr2806) - Transcr Unit
TU710 (ssr2843) - Transcr Unit
TU724 (ssr2848) - Transcr Unit
TU1516 (ssr2899) - Transcr Unit
TU214 (ssr2972) - Transcr Unit
TU246 (ssr2998, slr1794, slr1795, ssr3000) - Transcr Unit
TU532 (ssr3122) - Transcr Unit
TU1269 (ssr3184, slr1881, slr1882) - Transcr Unit
TU1272 (ssr3188) - Transcr Unit
TU1276 (ssr3189, slr1885) - Transcr Unit
TU2336 (ssr3304, slr1943, slr1944) - Transcr Unit
TU1845 (ssr3341, slr1970) - Transcr Unit
TU1490 (ssr3402, slr2004, slr1712) - Transcr Unit
TU567 (ssr3465) - Transcr Unit
TU1571 (ssr3532, slr2080, slr2081) - Transcr Unit
TU1665 (ssr3570, ssr3571, ssr3572, slr2107) - Transcr Unit
TU5025 (ssr5020, slr5021, slr5022) - Transcr Unit
TU6002 (ssr6003, slr6004, slr6005, slr6006, slr6007, slr6008, slr6009) - Transcr Unit
TU6018 (ssr6019) - Transcr Unit
TU6027 (ssr6030) - Transcr Unit
TU6044 (ssr6046, slr6047) - Transcr Unit
TU6047 (ssr6048, slr6049, slr6050, slr6051) - Transcr Unit
TU6073 (ssr6078) - Transcr Unit
TU6082 (ssr6089) - Transcr Unit
TU6093 (ssr6099, slr6100, slr6101, slr6102) - Transcr Unit
TU7029 (ssr7036) - Transcr Unit
TU7030 (ssr7036) - Transcr Unit
TU7034 (ssr7040, slr7041) - Transcr Unit
TU8009 (ssr8013, slr8014, slr8015, slr8016) - Transcr Unit
ssrA/tmRNA/ncl1680 (ncRNA) - TU3496
TU3496 (ssrA/tmRNA) - Transcr Unit
stpA/slr0746 (mRNA / CDS) - TU3586
stpA/slr0746 (Protein) - TU3586
sucC/sll1023 (mRNA / CDS) - TU374
sucC/sll1023 (Protein) - TU374
sucD/sll1557 (mRNA / CDS) - TU2049
sucD/sll1557 (Protein) - TU2049
sulA/slr1223 (mRNA / CDS) - TU1048
sulA/slr1223 (Protein) - TU1048
sycrp1/sll1371 (mRNA / CDS) - TU1922
sycrp1/sll1371 (Protein) - TU1922
TU1922 (sll1371) - Transcr Unit
TU762 (sll1924) - Transcr Unit
syR11/ncr1160 (ncRNA) - TU2440
TU2544 (SyR46, sll0315, sll0317) - Transcr Unit
syR47/ncr1080 (ncRNA) - TU2352
TU2351 (SyR47) - Transcr Unit
TU2352 (SyR47, slr0993) - Transcr Unit
syR48/ncl0830 (ncRNA) - TU1763
TU3300 (SyR5, sll0737, sll0738) - Transcr Unit
TU809 (SyR50) - Transcr Unit
TU810 (SyR50, slr1147) - Transcr Unit
TU1388 (SyR53) - Transcr Unit
syR6/ncl0880 (ncRNA) - TU1880
TU1880 (SyR6) - Transcr Unit
TU1881 (SyR6) - Transcr Unit
TU1804 (SyR7, sll1247) - Transcr Unit
syR8/ncl0030 (ncRNA) - TU99
TU99 (SyR8) - Transcr Unit
TU2629 (SyR9) - Transcr Unit
TU2628 (SyR9, sll0208) - Transcr Unit
talB/slr1793 (mRNA / CDS) - TU244
talB/slr1793 (Protein) - TU244
slr1793-3UTR (3'UTR) - TU244
slr1793-5UTR (5'UTR) - TU244
tatD/sll1786 (mRNA / CDS) - TU865
tatD/sll1786 (Protein) - TU865
tgt/slr0713 (mRNA / CDS) - TU3290
tgt/slr0713 (Protein) - TU3290
TU3290 (slr0713) - Transcr Unit
thiC/slr0118 (mRNA / CDS) - TU3165
thiC/slr0118 (Protein) - TU3165
slr0118-5UTR (5'UTR) - TU3165
TU3165 (slr0118, slr0119) - Transcr Unit
thiE/sll0635 (mRNA / CDS) - TU429
thiE/sll0635 (Protein) - TU429
thiG/slr0633 (mRNA / CDS) - TU2805
thiG/slr0633 (Protein) - TU2805
TU2805 (slr0633) - Transcr Unit
slr0633-3UTR (3'UTR) - TU2805
thrA/sll0455 (mRNA / CDS) - TU2774
thrA/sll0455 (Protein) - TU2774
TU2774 (sll0455) - Transcr Unit
thrB/sll1760 (mRNA / CDS) - TU1266
thrB/sll1760 (Protein) - TU1266
TU1266 (sll1760) - Transcr Unit
thrC/sll1172 (mRNA / CDS) - TU1952
thrC/sll1172 (Protein) - TU1952
TU1952 (sll1172) - Transcr Unit
thrC/sll1688 (mRNA / CDS) - TU1340
thrC/sll1688 (Protein) - TU1340
TU1340 (sll1688) - Transcr Unit
sll1688-3UTR (3'UTR) - TU1340
thrS/sll0078 (mRNA / CDS) - TU3050
thrS/sll0078 (Protein) - TU3050
TU3050 (sll0078) - Transcr Unit
thyX/sll1635 (mRNA / CDS) - TU530
thyX/sll1635 (Protein) - TU530
tig/sll0533 (mRNA / CDS) - TU3416
tig/sll0533 (Protein) - TU3416
sll0533-5UTR (5'UTR) - TU3416
tktA/sll1070 (mRNA / CDS) - TU819
tktA/sll1070 (Protein) - TU819
sll1070-3UTR (3'UTR) - TU819
sll1070-5UTR (5'UTR) - TU819
sll1070-as (asRNA)
tldD/slr1322 (mRNA / CDS) - TU1686
tldD/slr1322 (Protein) - TU1686
TU1686 (slr1322) - Transcr Unit
slr1322-3UTR (3'UTR) - TU1686
todF/sll1129 (mRNA / CDS) - TU1081
todF/sll1129 (Protein) - TU1081
topA/slr2058 (mRNA / CDS) - TU1455
topA/slr2058 (Protein) - TU1455
tpi/slr0783 (mRNA / CDS) - TU2522
tpi/slr0783 (Protein) - TU2522
TU2522 (slr0783, slr0784) - Transcr Unit
tpx, ycf42/sll0755 (mRNA / CDS) - TU2514
tpx, ycf42/sll0755 (Protein) - TU2514
trmB/sll1300 (mRNA / CDS) - TU673
trmB/sll1300 (Protein) - TU673
trmD/sll1198 (mRNA / CDS) - TU289
trmD/sll1198 (Protein) - TU289
trmE/sll1615 (mRNA / CDS)
trmE/sll1615 (Protein)
trpA/slr0966 (mRNA / CDS) - TU324
trpA/slr0966 (Protein) - TU324
TU324 (slr0966) - Transcr Unit
slr0966-3UTR (3'UTR) - TU324
slr0966-5UTR (5'UTR) - TU324
trpB/slr0543 (mRNA / CDS) - TU3385
trpB/slr0543 (Protein) - TU3385
trpC/slr0546 (mRNA / CDS) - TU3411
trpC/slr0546 (Protein) - TU3411
TU3411 (slr0546) - Transcr Unit
trpD/slr1867 (mRNA / CDS) - TU1235
trpD/slr1867 (Protein) - TU1235
TU1235 (slr1867) - Transcr Unit
slr1867-3UTR (3'UTR) - TU1235
trpE/slr0738 (mRNA / CDS) - TU113
trpE/slr0738 (Protein) - TU113
trpE/slr1979 (mRNA / CDS)
trpE/slr1979 (Protein)
trpG/slr0055 (mRNA / CDS) - TU2712
trpG/slr0055 (Protein) - TU2712
trpS/slr1884 (mRNA / CDS) - TU1275
trpS/slr1884 (Protein) - TU1275
TU1275 (slr1884) - Transcr Unit
slr1884-3UTR (3'UTR) - TU1275
truA/sll1820 (mRNA / CDS) - TU833
truA/sll1820 (Protein) - TU833
trxA/sll1980 (mRNA / CDS) - TU1597
trxA/sll1980 (Protein) - TU1597
TU1597 (sll1980) - Transcr Unit
sll1980-3UTR (3'UTR) - TU1597
trxA/slr0623 (mRNA / CDS) - TU3128
trxA/slr0623 (Protein) - TU3128
TU3128 (slr0623) - Transcr Unit
slr0623-3UTR (3'UTR) - TU3128
slr0623-5UTR (5'UTR) - TU3128
trxA/slr1139 (mRNA / CDS) - TU793
trxA/slr1139 (Protein) - TU793
TU793 (slr1139, slr1140) - Transcr Unit
trxM1/slr0233 (mRNA / CDS) - TU2661
trxM1/slr0233 (Protein) - TU2661
TU2661 (slr0233) - Transcr Unit
slr0233-3UTR (3'UTR) - TU2661
slr0233-5UTR (5'UTR) - TU2661
trxM2/sll1057 (mRNA / CDS) - TU73
trxM2/sll1057 (Protein) - TU73
TU73 (sll1057) - Transcr Unit
tsf/sll1261 (mRNA / CDS) - TU1777
tsf/sll1261 (Protein) - TU1777
TU1777 (sll1261) - Transcr Unit
sll1261-3UTR (3'UTR) - TU1777
tufA/sll1099 (mRNA / CDS) - TU1986
tufA/sll1099 (Protein) - TU1986
tyrA/slr2081 (mRNA / CDS) - TU1571
tyrA/slr2081 (Protein) - TU1571
tyrS/slr1031 (mRNA / CDS) - TU656
tyrS/slr1031,ssr1720 (Protein)
ubiX/slr1099 (mRNA / CDS) - TU186
ubiX/slr1099 (Protein) - TU186
upp/sll1035 (mRNA / CDS) - TU198
upp/sll1035 (Protein) - TU198
ureA/slr1256 (Protein) - TU1442
TU1442 (slr1256, 6803t14) - Transcr Unit
ureB/sll0420 (Protein) - TU2179
ureC/sll1750 (mRNA / CDS) - TU537
ureC/sll1750 (Protein) - TU537
TU520 (sll1639, sll1640, sll1641, ncl0190) - Transcr Unit
ureE/slr1219 (mRNA / CDS) - TU1037
ureE/slr1219 (Protein) - TU1037
TU858 (slr1899, slr1900, slr1901) - Transcr Unit
ureG/sll0643 (mRNA / CDS) - TU407
ureG/sll0643 (Protein) - TU407
TU407 (sll0643) - Transcr Unit
urtA/slr0447 (mRNA / CDS) - TU2184
urtA/slr0447 (Protein) - TU2184
urtD/sll0764 (mRNA / CDS) - TU2507
urtD/sll0764 (Protein) - TU2507
braG/sll0374 (mRNA / CDS) - TU2863
urtE/sll0374 (Protein) - TU2863
uvrA/slr1844 (mRNA / CDS) - TU974
uvrA/slr1844 (Protein) - TU974
uvrB/sll0459 (mRNA / CDS) - TU2750
uvrB/sll0459 (Protein) - TU2750
TU2749 (sll0459) - Transcr Unit
valS/slr0557 (mRNA / CDS) - TU3431
valS/slr0557 (Protein) - TU3431
TU3431 (slr0557) - Transcr Unit
vgb/sll0173 (mRNA / CDS) - TU2398
vgb/sll0173 (Protein) - TU2398
vipp1/sll0617 (mRNA / CDS) - TU2794
vipp1/sll0617 (Protein) - TU2794
TU2794 (sll0617) - Transcr Unit
sll0617-3UTR (3'UTR) - TU2794
sll0617-5UTR (5'UTR) - TU2794
xthA/sll1854 (mRNA / CDS)
xthA/sll1854 (Protein)
xylR/slr0329 (mRNA / CDS) - TU2370
xylR/slr0329 (Protein) - TU2370
TU3305 (sll0047) - Transcr Unit
ycf16/slr0075 (mRNA / CDS) - TU3030
ycf16/slr0075 (Protein) - TU3030
TU286 (ssr2142) - Transcr Unit
TU849 (sll1797) - Transcr Unit
ycf22/sll0751 (mRNA / CDS) - TU2516
ycf22/sll0751 (Protein) - TU2516
ycf22/sll1002 (mRNA / CDS) - TU503
ycf22/sll1002 (Protein) - TU503
TU2516 (sll0751, sll0752) - Transcr Unit
TU503 (sll1002, ssl1920) - Transcr Unit
ycf23/slr2032 (mRNA / CDS) - TU541
ycf23/slr2032 (Protein) - TU541
TU541 (slr2032) - Transcr Unit
slr2032-3UTR (3'UTR) - TU541
slr2032-5UTR (5'UTR) - TU541
ycf24/slr0074 (mRNA / CDS) - TU3030
ycf24/slr0074 (Protein) - TU3030
slr0074-5UTR (5'UTR) - TU3030
TU3030 (slr0074, slr0075, slr0076, slr0077) - Transcr Unit
ycf29/slr1783 (mRNA / CDS)
ycf29/slr1783 (Protein)
ycf3/slr0823 (mRNA / CDS) - TU1771
ycf3/slr0823 (Protein) - TU1771
TU1771 (slr0823) - Transcr Unit
slr0823-3UTR (3'UTR) - TU1771
ycf30/sll0998 (mRNA / CDS) - TU626
ycf30/sll0998 (Protein) - TU626
TU626 (sll0998) - Transcr Unit
sll0998-3UTR (3'UTR) - TU626
sll0998-5UTR (5'UTR) - TU626
TU3011 (ssr1425) - Transcr Unit
ycf35/sll0661 (mRNA / CDS) - TU3286
ycf35/sll0661 (Protein) - TU3286
TU3130 (sll0584) - Transcr Unit
ycf37/slr0171 (Protein) - TU2394
ycf38/sll0760 (mRNA / CDS) - TU2508
ycf38/sll0760 (Protein) - TU2508
TU2508 (sll0760, sll0761, sll0762, sll0763, ncl1210) - Transcr Unit
ycf39/sll1218 (mRNA / CDS) - TU1720
ycf39/sll1218 (Protein) - TU1720
ycf39/slr0399 (mRNA / CDS) - TU2227
ycf39/slr0399 (Protein) - TU2227
TU2227 (slr0399) - Transcr Unit
slr1218 (mRNA / CDS) - TU1037
ycf39/slr1218 (Protein) - TU1037
TU1037 (slr1218, slr1219) - Transcr Unit
ycf4/sll0226 (mRNA / CDS) - TU136
ycf4/sll0226 (Protein) - TU136
sll0226-5UTR (5'UTR) - TU136
TU136 (sll0226, sll0227, sll0228) - Transcr Unit
ycf40/ssr0102 (Protein) - TU2696
ycf41/slr1034 (mRNA / CDS) - TU659
ycf41/slr1034 (Protein) - TU659
slr1034-as (asRNA)
TU659 (slr1034, slr1035) - Transcr Unit
TU2656 (sll0194) - Transcr Unit
ycf45/slr0692 (mRNA / CDS) - TU428
ycf45/slr0692 (Protein) - TU428
TU428 (slr0692) - Transcr Unit
slr0692-3UTR (3'UTR) - TU428
ycf46/slr0480 (mRNA / CDS) - TU2755
ycf46/slr0480 (Protein) - TU2755
ycf48/slr2034 (mRNA / CDS) - TU545
ycf48/slr2034 (Protein) - TU545
TU2812 (sll0608, sll0609) - Transcr Unit
ycf50/slr2073 (mRNA / CDS) - TU919
ycf50/slr2073 (Protein) - TU919
TU919 (slr2073) - Transcr Unit
slr2073-3UTR (3'UTR) - TU919
slr2073-5UTR (5'UTR) - TU919
ycf51/sll1702 (mRNA / CDS) - TU1318
ycf51/sll1702 (Protein) - TU1318
TU1318 (sll1702, sll1703) - Transcr Unit
sll0286 (mRNA / CDS) - TU3191
ycf52/sll0286 (Protein) - TU3191
TU3191 (sll0286) - Transcr Unit
ycf53/sll0558 (mRNA / CDS) - TU5
ycf53/sll0558 (Protein) - TU5
TU5 (sll0558) - Transcr Unit
ycf54/slr1780 (Protein) - TU224
ycf55/sll1879 (mRNA / CDS) - TU1866
ycf55/sll1879 (Protein) - TU1866
TU1866 (sll1879) - Transcr Unit
ycf56/slr0050 (mRNA / CDS) - TU2696
ycf56/slr0050 (Protein) - TU2696
ycf57/slr1417 (mRNA / CDS) - TU176
ycf57/slr1417 (Protein) - TU176
TU176 (slr1417) - Transcr Unit
slr1417-3UTR (3'UTR) - TU176
slr1417-5UTR (5'UTR) - TU176
ycf58/slr2049 (mRNA / CDS) - TU582
ycf58/slr2049 (Protein) - TU582
ycf59/sll1214 (mRNA / CDS) - TU8
ycf59/sll1214 (Protein) - TU8
TU8 (sll1214) - Transcr Unit
sll1214-3UTR (3'UTR) - TU8
sll1214-5UTR (5'UTR) - TU8
ycf60/sll1737 (mRNA / CDS) - TU941
ycf60/sll1737 (Protein) - TU941
sll1737-5UTR (5'UTR) - TU941
TU941 (sll1737, sll1738, ncl0420) - Transcr Unit
rpoZ/ycf61/ssl2982 (Protein) - TU2128
TU2128 (ssl2982, sll1532) - Transcr Unit
ycf64/slr1846 (mRNA / CDS) - TU533
ycf64/slr1846 (Protein) - TU533
TU533 (slr1846) - Transcr Unit
slr1846-3UTR (3'UTR) - TU533
ycf65/slr0923 (mRNA / CDS) - TU2943
ycf65/slr0923 (Protein) - TU2943
TU2943 (slr0923, ncr1440) - Transcr Unit
ycf66/slr0503 (mRNA / CDS) - TU3072
ycf66/slr0503 (Protein) - TU3072
TU3072 (slr0503, slr0505) - Transcr Unit
ycf81/slr1972 (mRNA / CDS) - TU1852
ycf81/slr1972 (Protein) - TU1852
ycf84/slr0882 (mRNA / CDS) - TU1182
ycf84/slr0882 (Protein) - TU1182
TU1182 (slr0882) - Transcr Unit
slr0882-as (asRNA)
ycf85/slr0251 (mRNA / CDS) - TU142
ycf85/slr0251 (Protein) - TU142
TU142 (slr0251, slr0252) - Transcr Unit
yefA/slr1072 (mRNA / CDS) - TU355
yefA/slr1072 (Protein) - TU355
Yfr1/ncr1525 (ncRNA) - TU3126
TU3125 (Yfr1) - Transcr Unit
TU3126 (Yfr1) - Transcr Unit
yfr2a/ncl0720 (ncRNA) - TU1596
TU1596 (yfr2a) - Transcr Unit
yfr2b/ncr1397 (ncRNA) - TU2885
TU2885 (yfr2b, slr0199) - Transcr Unit
yfr2c/ncl1725 (ncRNA) - TU3575
TU3575 (yfr2c, sll1477) - Transcr Unit
yibO/slr1945 (mRNA / CDS) - TU2338
yibO/slr1945 (Protein) - TU2338
slr1945-5UTR (5'UTR) - TU2338
ymxG/slr1331 (mRNA / CDS) - TU1711
ymxG/slr1331 (Protein) - TU1711
ytfC/slr1761 (mRNA / CDS) - TU514
ytfC/slr1761 (Protein) - TU514
slr1761-3UTR (3'UTR) - TU514
zam/sll1910 (mRNA / CDS) - TU784
zam/sll1910 (Protein) - TU784
ziaR/sll0792 (Protein) - TU3225
zntC/slr2043 (mRNA / CDS) - TU570
zntC/slr2043 (Protein) - TU570
zwf/slr1843 (mRNA / CDS) - TU974
zwf/slr1843 (Protein) - TU974
TU974 (slr1843, slr1844) - Transcr Unit
6803t01-3UTR (3'UTR)
6803t02-5UTR (5'UTR) - TU241
6803t03-3UTR (3'UTR) - TU256
6803t03-5UTR (5'UTR) - TU256
6803t04-5UTR (5'UTR) - TU317
6803t06-5UTR (5'UTR) - TU430
6803t07-3UTR (3'UTR) - TU629
6803t07-5UTR (5'UTR) - TU629
6803t08-5UTR (5'UTR) - TU641
6803t10-3UTR (3'UTR) - TU1004
6803t10-5UTR (5'UTR) - TU1004
6803t11-3UTR (3'UTR) - TU1111
6803t11-5UTR (5'UTR) - TU1111
6803t15-3UTR (3'UTR) - TU1587
6803t15-5UTR (5'UTR) - TU1587
6803t16-5UTR (5'UTR) - TU1663
6803t18-3UTR (3'UTR) - TU1780
6803t18-5UTR (5'UTR) - TU1780
6803t19-3UTR (3'UTR) - TU1890
6803t19-5UTR (5'UTR) - TU1890
6803t20-3UTR (3'UTR) - TU1904
6803t21-3UTR (3'UTR)
6803t22-3UTR (3'UTR) - TU2132
6803t22-5UTR (5'UTR) - TU2132
6803t24-3UTR (3'UTR)
6803t25-3UTR (3'UTR)
6803t26-3UTR (3'UTR)
6803t31-3UTR (3'UTR) - TU2613
6803t32-3UTR (3'UTR) - TU2719
6803t32-5UTR (5'UTR) - TU2719
6803t33-3UTR (3'UTR) - TU2792
6803t33-5UTR (5'UTR) - TU2792
6803t34-5UTR (5'UTR) - TU2935
6803t37-5UTR (5'UTR) - TU3394
6803t39-3UTR (3'UTR) - TU3465
6803t39-5UTR (5'UTR) - TU3465
6803t41-3UTR (3'UTR) - TU3514
6803t41-5UTR (5'UTR) - TU3514
6803t42-3UTR (3'UTR)
cr1RNA (CRISPR RNA)
cr2RNA (CRISPR RNA)
cr3RNA (CRISPR RNA)
hliR1 (mRNA / CDS) - TU1647
ncl0040-3UTR (3'UTR) - TU123
ncl0160-3UTR (3'UTR) - TU475
ncl0160-5UTR (5'UTR) - TU475
ncl0270-3UTR (3'UTR) - TU715
ncl0270-5UTR (5'UTR) - TU715
ncl0390-5UTR (5'UTR) - TU853
ncl0590-3UTR (3'UTR)
ncl0620-3UTR (3'UTR) - TU1440
ncl0620-5UTR (5'UTR) - TU1440
ncl0640-3UTR (3'UTR) - TU1470
ncl0720-3UTR (3'UTR)
ncl0850-5UTR (5'UTR) - TU1825
ncl0950-3UTR (3'UTR) - TU1997
ncl1020-3UTR (3'UTR) - TU2142
ncl1020-5UTR (5'UTR) - TU2142
ncl1060-3UTR (3'UTR) - TU2206
ncl1090-3UTR (3'UTR) - TU2340
ncl1130-3UTR (3'UTR) - TU2366
ncl1330-3UTR (3'UTR) - TU2824
ncl1350-3UTR (3'UTR) - TU2857
ncl1680-3UTR (3'UTR)
ncl1720-3UTR (3'UTR) - TU3536
ncl1720-5UTR (5'UTR) - TU3536
ncl1790-3UTR (3'UTR)
ncr0100-3UTR (3'UTR) - TU206
ncr0200-5UTR (5'UTR) - TU420
ncr0310-5UTR (5'UTR) - TU731
ncr0570-3UTR (3'UTR) - TU1471
ncr0570-5UTR (5'UTR) - TU1471
ncr0670-3UTR (3'UTR) - TU1695
ncr0870-3UTR (3'UTR)
ncr0910-3UTR (3'UTR)
ncr1100-3UTR (3'UTR) - TU2383
ncr1160-3UTR (3'UTR) - TU2440
ncr1160-5UTR (5'UTR) - TU2440
ncr1220-5UTR (5'UTR) - TU2529
ncr1525-3UTR (3'UTR)
ncr1640-5UTR (5'UTR) - TU3492
ncr1690-3UTR (3'UTR) - TU3604
Norf4 (mRNA / CDS) - TU1188
Norf4-3UTR (3'UTR) - TU1188
Norf4-5UTR (5'UTR) - TU1188
Norf5 (mRNA / CDS) - TU1457
Norf6 (mRNA / CDS) - TU1918
Norf6-5UTR (5'UTR) - TU1918
nsiR6 (mRNA / CDS)
Parent=desC%2Cdes9 (mRNA / CDS)
sgl0002 (mRNA / CDS) - TU149
sll0024 (mRNA / CDS) - TU2567
sll0031 (mRNA / CDS) - TU3328
ycf12/sll0047 (mRNA / CDS) - TU3305
sll0047-3UTR (3'UTR) - TU3305
sll0047-5UTR (5'UTR) - TU3305
sll0060 (mRNA / CDS) - TU2721
sll0062 (mRNA / CDS) - TU2716
sll0156 (mRNA / CDS) - TU2427
sll0156-5UTR (5'UTR) - TU2427
sll0168 (mRNA / CDS) - TU2408
sll0189 (mRNA / CDS) - TU2874
sll0191 (mRNA / CDS) - TU2658
sll0191-5UTR (5'UTR) - TU2658
ycf43/sll0194 (mRNA / CDS) - TU2656
sll0195 (mRNA / CDS) - TU2654
sll0210 (mRNA / CDS) - TU2617
phoA/sll0222 (mRNA / CDS) - TU148
sll0222-5UTR (5'UTR) - TU148
sll0225 (mRNA / CDS) - TU137
sll0238 (mRNA / CDS) - TU1569
sll0240 (mRNA / CDS) - TU1569
sll0241 (mRNA / CDS) - TU1569
isiA/sll0247 (mRNA / CDS) - TU1556
sll0247-5UTR (5'UTR) - TU1556
sll0247-as (asRNA) - TU1557
sll0247-as2 (asRNA) - TU1557
sll0252 (mRNA / CDS) - TU1543
tsaE/sll0257 (mRNA / CDS)
sll0261 (mRNA / CDS) - TU2202
desD(des6)/sll0262 (mRNA / CDS) - TU2196
sll0264 (mRNA / CDS) - TU2194
sll0266 (mRNA / CDS) - TU2194
priA/sll0270 (mRNA / CDS) - TU2190
sll0294 (mRNA / CDS) - TU2388
sll0297 (mRNA / CDS) - TU2371
sll0297-5UTR (5'UTR) - TU2371
sll0298 (mRNA / CDS) - TU2371
hypF/sll0322 (mRNA / CDS) - TU2532
sll0327 (mRNA / CDS) - TU2481
fabG/sll0330 (mRNA / CDS) - TU2474
sll0350 (mRNA / CDS) - TU2441
sll0355 (mRNA / CDS) - TU2237
sll0360 (mRNA / CDS) - TU2223
sll0360-5UTR (5'UTR) - TU2223
sll0371 (mRNA / CDS) - TU2867
sll0372 (mRNA / CDS) - TU2863
sll0375 (mRNA / CDS)
sll0375-as (asRNA)
mfd/sll0377 (mRNA / CDS) - TU2854
cobA/sll0378 (mRNA / CDS) - TU2852
sll0378-5UTR (5'UTR) - TU2852
sll0382 (mRNA / CDS) - TU2838
cbiM/sll0383 (mRNA / CDS) - TU2838
sll0384 (mRNA / CDS) - TU2838
sll0385 (mRNA / CDS) - TU2838
sll0397 (mRNA / CDS) - TU2831
sll0423 (mRNA / CDS) - TU2174
sll0423-5UTR (5'UTR) - TU2174
sll0424 (mRNA / CDS) - TU2173
sll0442 (mRNA / CDS)
sll0448 (mRNA / CDS)
sll0473 (mRNA / CDS) - TU3094
sll0473-5UTR (5'UTR) - TU3094
hik28/sll0474 (mRNA / CDS) - TU3091
sll0477 (mRNA / CDS) - TU3083
sll0478 (mRNA / CDS) - TU3083
sll0481 (mRNA / CDS) - TU3075
sll0481-5UTR (5'UTR) - TU3075
sll0484 (mRNA / CDS) - TU3068
sll0494 (mRNA / CDS) - TU3396
sll0496 (mRNA / CDS) - TU3393
sll0496-5UTR (5'UTR) - TU3393
sll0503 (mRNA / CDS) - TU3381
sll0536 (mRNA / CDS) - TU3412
amt3/sll0537 (mRNA / CDS) - TU3412
sll0539 (mRNA / CDS) - TU2980
sll0539-5UTR (5'UTR) - TU2980
sll0543 (mRNA / CDS) - TU2962
sll0549 (mRNA / CDS) - TU2955
sll0552 (mRNA / CDS) - TU2951
map/sll0555 (mRNA / CDS) - TU2948
arcC/sll0573 (mRNA / CDS)
ycf36/sll0584 (mRNA / CDS) - TU3130
cysR/sll0594 (mRNA / CDS) - TU3107
ycf49/sll0608 (mRNA / CDS) - TU2812
ruvB/sll0613 (mRNA / CDS) - TU2803
ccdA/sll0621 (mRNA / CDS) - TU3528
sll0621-5UTR (5'UTR) - TU3528
lim/sll0648 (mRNA / CDS) - TU403
sll0664 (mRNA / CDS) - TU3281
pstC/sll0681 (mRNA / CDS) - TU2785
sll0686 (mRNA / CDS) - TU2784
sigI/sll0687 (mRNA / CDS) - TU2783
sll0687-5UTR (5'UTR) - TU2783
sll0688 (mRNA / CDS) - TU2782
sll0696 (mRNA / CDS)
nifS/sll0704 (mRNA / CDS) - TU108
sll0704-3UTR (3'UTR) - TU108
sll0704-5UTR (5'UTR) - TU108
sll0721 (mRNA / CDS) - TU3621
sll0722 (mRNA / CDS) - TU3620
sll0727 (mRNA / CDS) - TU3609
modA/sll0738 (mRNA / CDS) - TU3300
sll0739 (mRNA / CDS)
sll0742 (mRNA / CDS) - TU3296
sll0763 (mRNA / CDS) - TU2508
sll0765 (mRNA / CDS) - TU2503
sll0765-as (asRNA)
radC/sll0766 (mRNA / CDS) - TU2503
sll0775 (mRNA / CDS) - TU3253
sll0778 (mRNA / CDS) - TU3252
merR/sll0784 (mRNA / CDS) - TU3241
sll0787 (mRNA / CDS)
sll0793 (mRNA / CDS) - TU3224
corR, coaR/sll0794 (mRNA / CDS) - TU3222
sll0797 (mRNA / CDS) - TU3215
sll0798 (mRNA / CDS)
sll0802 (mRNA / CDS) - TU1773
sll0810 (mRNA / CDS) - TU1750
sll0812 (mRNA / CDS) - TU1747
rsmI/sll0818 (mRNA / CDS) - TU1735
pcnB/sll0825 (mRNA / CDS) - TU3010
sll0843 (mRNA / CDS) - TU1398
sll0843-5UTR (5'UTR) - TU1398
sll0847 (mRNA / CDS) - TU1389
dnaA/sll0848 (mRNA / CDS)
sigH/sll0856 (mRNA / CDS) - TU1380
sll0856-5UTR (5'UTR) - TU1380
sll0857 (mRNA / CDS) - TU1380
sll0864 (mRNA / CDS)
rsgA/sll0898 (mRNA / CDS) - TU3409
sll0898-as (asRNA)
sll0910 (mRNA / CDS) - TU2090
sll0912 (mRNA / CDS) - TU2087
sll0922-as (asRNA)
sll0924 (mRNA / CDS) - TU343
sll0924-5UTR (5'UTR) - TU343
dus2/sll0926 (mRNA / CDS) - TU337
sll0930 (mRNA / CDS) - TU333
sll0932 (mRNA / CDS) - TU333
sll0983 (mRNA / CDS) - TU664
sll0992 (mRNA / CDS) - TU641
sll1006 (mRNA / CDS) - TU498
sll1011 (mRNA / CDS) - TU487
sll1024 (mRNA / CDS) - TU374
sll1037 (mRNA / CDS) - TU198
sll1040 (mRNA / CDS) - TU178
hypA2/sll1078 (mRNA / CDS) - TU795
sll1086 (mRNA / CDS) - TU2013
sll1086-3UTR (3'UTR) - TU2013
sll1086-5UTR (5'UTR) - TU2013
gtrA/sll1102 (mRNA / CDS) - TU1985
sll1119 (mRNA / CDS) - TU879
sll1119-3UTR (3'UTR) - TU879
sll1119-5UTR (5'UTR) - TU879
sll1131 (mRNA / CDS) - TU1074
sll1132 (mRNA / CDS) - TU1073
mraW/sll1144 (mRNA / CDS) - TU1036
norA/sll1154 (mRNA / CDS) - TU1427
sll1161 (mRNA / CDS) - TU1420
sll1164 (mRNA / CDS) - TU1420
sll1169 (mRNA / CDS)
sll1170 (mRNA / CDS)
lgt/sll1187 (mRNA / CDS) - TU310
sll1191 (mRNA / CDS) - TU297
sll1191-as (asRNA)
crcB/sll1192 (mRNA / CDS) - TU297
sll1202 (mRNA / CDS)
sll1203 (mRNA / CDS)
sll1204 (mRNA / CDS)
pchR/sll1205 (mRNA / CDS)
iutA/sll1206 (mRNA / CDS) - TU24
sll1219 (mRNA / CDS) - TU1719
sll1225 (mRNA / CDS) - TU1714
cytM/sll1245 (mRNA / CDS) - TU1807
sll1245-3UTR (3'UTR) - TU1807
recF/sll1277 (mRNA / CDS) - TU1131
psbZ, ycf9/sll1281 (mRNA / CDS) - TU1125
sll1283 (mRNA / CDS) - TU702
atp1/sll1321 (mRNA / CDS) - TU166
sll1321-5UTR (5'UTR) - TU166
sll1321-as (asRNA)
sll1333 (mRNA / CDS) - TU3466
sll1344 (mRNA / CDS) - TU3440
sll1344-3UTR (3'UTR) - TU3440
queG/sll1348 (mRNA / CDS) - TU1125
cbbZp/sll1349 (mRNA / CDS) - TU1124
hik15/sll1353 (mRNA / CDS) - TU1115
sll1373 (mRNA / CDS) - TU1921
sll1378 (mRNA / CDS) - TU1911
pppA/sll1387 (mRNA / CDS) - TU1892
sll1387-5UTR (5'UTR) - TU1892
sll1389 (mRNA / CDS) - TU1889
sll1392 (mRNA / CDS) - TU54
fhuA/sll1406 (mRNA / CDS) - TU32
fhuA/sll1409 (mRNA / CDS) - TU31
sll1428 (mRNA / CDS) - TU1618
desB/sll1441 (mRNA / CDS) - TU1967
crtR/sll1468 (mRNA / CDS) - TU998
sll1476 (mRNA / CDS)
sll1476-as (asRNA)
sll1482 (mRNA / CDS) - TU3572
sll1485 (mRNA / CDS) - TU3570
sll1485-5UTR (5'UTR) - TU3570
sll1488 (mRNA / CDS) - TU3559
sll1488-5UTR (5'UTR) - TU3559
sll1496 (mRNA / CDS) - TU3543
sll1496-5UTR (5'UTR) - TU3543
sll1500 (mRNA / CDS)
cobB/sll1501 (mRNA / CDS)
sll1503 (mRNA / CDS) - TU463
sll1511 (mRNA / CDS)
rfbP/sll1535 (mRNA / CDS) - TU2123
sll1535-3UTR (3'UTR) - TU2123
sll1535-5UTR (5'UTR) - TU2123
dpm1/sll1540 (mRNA / CDS)
sll1540-as (asRNA)
sll1544 (mRNA / CDS) - TU3653
sll1547 (mRNA / CDS) - TU3642
sll1550 (mRNA / CDS) - TU3630
dnaE/sll1572 (mRNA / CDS) - TU742
sll1572-as (asRNA)
sll1586 (mRNA / CDS) - TU1531
sll1586-as (asRNA)
hik20/sll1590 (mRNA / CDS) - TU1525
mntA/sll1599 (mRNA / CDS)
sll1606 (mRNA / CDS) - TU1503
sll1608 (mRNA / CDS) - TU1497
sll1609 (mRNA / CDS) - TU1497
sll1611 (mRNA / CDS) - TU1496
phr/sll1629 (mRNA / CDS) - TU1347
sll1632 (mRNA / CDS) - TU1033
ureD/sll1639 (mRNA / CDS) - TU520
sll1639-5UTR (5'UTR) - TU520
sll1642 (mRNA / CDS) - TU519
hrcA/sll1670 (mRNA / CDS) - TU241
sll1681 (mRNA / CDS) - TU225
sll1681-as (asRNA)
ycf10/sll1685 (mRNA / CDS)
sll1686 (mRNA / CDS) - TU213
sll1717 (mRNA / CDS) - TU978
sll1722 (mRNA / CDS) - TU962
sll1722-5UTR (5'UTR) - TU962
icsA/sll1724 (mRNA / CDS)
sll1726 (mRNA / CDS)
sll1736 (mRNA / CDS) - TU948
sll1736-3UTR (3'UTR) - TU948
sll1736-5UTR (5'UTR) - TU948
sll1738 (mRNA / CDS) - TU941
sll1752 (mRNA / CDS) - TU531
sll1752-5UTR (5'UTR) - TU531
sll1755 (mRNA / CDS) - TU1280
sll1755-as2 (asRNA)
glmM/sll1758 (mRNA / CDS) - TU1267
sll1758-3UTR (3'UTR) - TU1267
sll1758-5UTR (5'UTR) - TU1267
sll1761 (mRNA / CDS)
sll1763 (mRNA / CDS) - TU1263
sll1763-3UTR (3'UTR) - TU1263
sll1765 (mRNA / CDS) - TU1262
mutS/sll1772 (mRNA / CDS) - TU1249
sll1774 (mRNA / CDS) - TU1241
ycf21/sll1797 (mRNA / CDS) - TU849
sll1832 (mRNA / CDS) - TU616
sll1832-5UTR (5'UTR) - TU616
ftsI/sll1833 (mRNA / CDS) - TU616
sll1845 (mRNA / CDS) - TU2332
sll1849 (mRNA / CDS) - TU2322
sll1853 (mRNA / CDS) - TU2319
sll1858 (mRNA / CDS) - TU2315
sll1858-5UTR (5'UTR) - TU2315
sll1861-5UTR (5'UTR) - TU2309
sll1864 (mRNA / CDS)
dnaG/sll1868 (mRNA / CDS)
lytR/sll1872 (mRNA / CDS) - TU1873
ho2/sll1875 (mRNA / CDS)
cobN/sll1890 (mRNA / CDS) - TU1837
sll1898 (mRNA / CDS) - TU1818
ctaB/sll1899 (mRNA / CDS) - TU1818
hemN/sll1917 (mRNA / CDS)
sycrp2/sll1924 (mRNA / CDS) - TU762
comE/sll1929 (mRNA / CDS) - TU595
sll1930-5UTR (5'UTR) - TU593
sll1934-3UTR (3'UTR) - TU586
gyrA/sll1941 (mRNA / CDS) - TU560
sll1951 (mRNA / CDS) - TU1463
sll1951-5UTR (5'UTR) - TU1463
sll1954 (mRNA / CDS)
sll1960 (mRNA / CDS) - TU1450
pmgA/sll1968 (mRNA / CDS) - TU914
sll1968-5UTR (5'UTR) - TU914
sll1969 (mRNA / CDS) - TU914
sll1982-3UTR (3'UTR) - TU1590
petL/sll1994a (mRNA / CDS)
sll1995 (mRNA / CDS) - TU1681
sll1997-5UTR (5'UTR) - TU1669
sll2007 (mRNA / CDS) - TU1298
sigD/sll2012 (mRNA / CDS) - TU1288
sll2013 (mRNA / CDS) - TU1288
sll5014 (mRNA / CDS)
sll5026 (mRNA / CDS) - TU5029
sll5028 (mRNA / CDS) - TU5031
sll5028-5UTR (5'UTR) - TU5031
sll5030 (mRNA / CDS)
sll5032 (mRNA / CDS) - TU5036
sll5036 (mRNA / CDS) - TU5036
sll5041 (mRNA / CDS) - TU5046
sll5043 (mRNA / CDS) - TU5051
sll5046 (mRNA / CDS) - TU5051
sll5047 (mRNA / CDS) - TU5053
sll5049 (mRNA / CDS) - TU5056
sll5057 (mRNA / CDS) - TU5064
sll5057-3UTR (3'UTR) - TU5064
sll5063 (mRNA / CDS) - TU5077
sll5063-5UTR (5'UTR) - TU5077
sll5069 (mRNA / CDS) - TU5082
sll5069-5UTR (5'UTR) - TU5082
sll5076-5UTR (5'UTR) - TU5089
sll5083 (mRNA / CDS) - TU5096
sll5086 (mRNA / CDS)
sll5089 (mRNA / CDS)
sll5090 (mRNA / CDS)
sll5097 (mRNA / CDS) - TU5102
sll5109 (mRNA / CDS) - TU5111
sll6052 (mRNA / CDS)
sll6053 (mRNA / CDS) - TU6049
sll6059 (mRNA / CDS) - TU6055
sll6098 (mRNA / CDS) - TU6092
sll6098-3UTR (3'UTR) - TU6092
sll6098-5UTR (5'UTR) - TU6092
sll6109 (mRNA / CDS) - TU6101
sll7009 (mRNA / CDS)
sll7027-5UTR (5'UTR) - TU7015
sll7028 (mRNA / CDS)
sll7029 (mRNA / CDS) - TU7018
sll7047 (mRNA / CDS) - TU7043
sll7047-3UTR (3'UTR) - TU7043
sll7050 (mRNA / CDS) - TU7047
sll7050-3UTR (3'UTR) - TU7047
sll7055 (mRNA / CDS) - TU7051
sll7056-5UTR (5'UTR) - TU7052
sll7063 (mRNA / CDS) - TU7058
sll7064 (mRNA / CDS) - TU7058
sll7065 (mRNA / CDS) - TU7058
sll7066 (mRNA / CDS) - TU7058
sll7067 (mRNA / CDS) - TU7058
sll7075 (mRNA / CDS) - TU7071
sll7075-5UTR (5'UTR) - TU7071
sll8006 (mRNA / CDS) - TU8005
sll8009 (mRNA / CDS)
sll8017 (mRNA / CDS) - TU8012
sll8018 (mRNA / CDS) - TU8016
sll8018-5UTR (5'UTR) - TU8016
sll8019 (mRNA / CDS) - TU8019
sll8019-5UTR (5'UTR) - TU8019
sll8034 (mRNA / CDS)
sll8040 (mRNA / CDS)
sll8048-3UTR (3'UTR) - TU8043
sll8048-5UTR (5'UTR) - TU8043
sllt43 (mRNA / CDS)
mgtC/slr0014 (mRNA / CDS) - TU2588
slr0016 (mRNA / CDS) - TU2595
slr0019 (mRNA / CDS) - TU2601
recG/slr0020 (mRNA / CDS) - TU2607
aspC/slr0036 (mRNA / CDS)
slr0053 (mRNA / CDS) - TU2711
dgkA/slr0054 (mRNA / CDS) - TU2712
slr0054-5UTR (5'UTR) - TU2712
slr0059-5UTR (5'UTR) - TU2718
slr0081 (mRNA / CDS) - TU3043
slr0090-5UTR (5'UTR) - TU3057
slr0091 (mRNA / CDS)
slr0092 (mRNA / CDS)
slr0095 (mRNA / CDS) - TU3059
slr0096 (mRNA / CDS) - TU3060
slr0103 (mRNA / CDS)
slr0104 (mRNA / CDS) - TU3145
slr0104-5UTR (5'UTR) - TU3145
slr0109 (mRNA / CDS)
hat/slr0143 (mRNA / CDS) - TU2257
slr0152 (mRNA / CDS) - TU2267
slr0179 (mRNA / CDS) - TU2414
recO/slr0181 (mRNA / CDS) - TU2419
slr0195 (mRNA / CDS) - TU2880
slr0214 (mRNA / CDS) - TU2615
slr0224 (mRNA / CDS)
slr0232 (mRNA / CDS) - TU2660
slr0236 (mRNA / CDS) - TU119
slr0240 (mRNA / CDS) - TU126
slr0241 (mRNA / CDS) - TU126
cbiJ/slr0252 (mRNA / CDS) - TU142
slr0264 (mRNA / CDS) - TU1546
slr0265-5UTR (5'UTR) - TU1549
slr0271 (mRNA / CDS) - TU1565
slr0272 (mRNA / CDS) - TU1565
slr0273 (mRNA / CDS) - TU1565
slr0273-as (asRNA)
slr0284 (mRNA / CDS) - TU2203
slr0284-3UTR (3'UTR) - TU2203
slr0284-5UTR (5'UTR) - TU2203
slr0292 (mRNA / CDS) - TU2212
slr0292-5UTR (5'UTR) - TU2212
slr0294 (mRNA / CDS) - TU2215
slr0299 (mRNA / CDS) - TU3174
slr0304 (mRNA / CDS)
slr0305 (mRNA / CDS)
hik29/slr0311 (mRNA / CDS)
slr0314 (mRNA / CDS)
slr0316 (mRNA / CDS) - TU3202
blaOXA-3/slr0319 (mRNA / CDS)
slr0324 (mRNA / CDS) - TU2368
rfbW/slr0344 (mRNA / CDS)
slr0347 (mRNA / CDS) - TU2534
slr0360 (mRNA / CDS) - TU2435
slr0363 (mRNA / CDS) - TU2444
slr0364 (mRNA / CDS) - TU2447
slr0366 (mRNA / CDS) - TU2451
slr0368 (mRNA / CDS) - TU2453
slr0368-5UTR (5'UTR) - TU2453
slr0383 (mRNA / CDS) - TU2476
ntcB/slr0395 (mRNA / CDS) - TU2218
slr0421 (mRNA / CDS) - TU2849
rlpA/slr0423 (mRNA / CDS) - TU2859
slr0423-5UTR (5'UTR) - TU2859
slr0438 (mRNA / CDS) - TU2683
dnaX/slr0446 (mRNA / CDS) - TU2182
radA/slr0448 (mRNA / CDS) - TU2185
slr0449 (mRNA / CDS)
slr0456 (mRNA / CDS) - TU3675
truB/slr0457 (mRNA / CDS)
slr0468 (mRNA / CDS) - TU2739
murJ/slr0488 (mRNA / CDS) - TU2767
menE/slr0492 (mRNA / CDS) - TU2770
slr0495 (mRNA / CDS) - TU2775
slr0517 (mRNA / CDS) - TU3095
slr0517-as (asRNA)
abfB/slr0518 (mRNA / CDS) - TU3096
slr0518-3UTR (3'UTR) - TU3096
slr0518-5UTR (5'UTR) - TU3096
slr0521 (mRNA / CDS) - TU3097
slr0530 (mRNA / CDS)
slr0531 (mRNA / CDS)
hik10/slr0533 (mRNA / CDS) - TU3355
slr0533-5UTR (5'UTR) - TU3355
slr0554 (mRNA / CDS) - TU3420
slr0572-5UTR (5'UTR) - TU2961
slr0579 (mRNA / CDS) - TU2969
slr0587 (mRNA / CDS) - TU3690
slr0587-3UTR (3'UTR) - TU3690
slr0587-5UTR (5'UTR) - TU3690
slr0610 (mRNA / CDS) - TU3717
slr0612 (mRNA / CDS) - TU2
slr0612-as (asRNA)
slr0616 (mRNA / CDS) - TU3112
cobS/slr0636 (mRNA / CDS)
slr0639 (mRNA / CDS) - TU2811
slr0642 (mRNA / CDS) - TU2813
dus1/slr0644 (mRNA / CDS) - TU2818
slr0655 (mRNA / CDS) - TU3510
slr0655-as (asRNA)
slr0656 (mRNA / CDS) - TU3512
slr0656-3UTR (3'UTR) - TU3512
fmu/fmv/slr0679 (mRNA / CDS) - TU393
pleD/slr0687 (mRNA / CDS) - TU409
slr0700 (mRNA / CDS) - TU3256
merR/slr0701 (mRNA / CDS)
slr0702 (mRNA / CDS) - TU3257
slr0712 (mRNA / CDS) - TU3285
xerC/slr0733 (mRNA / CDS) - TU110
slr0743a (mRNA / CDS) - TU119
slr0748 (mRNA / CDS) - TU3592
chlL/slr0749 (mRNA / CDS) - TU3594
p/slr0753 (mRNA / CDS) - TU3599
slr0779 (mRNA / CDS) - TU2511
slr0779-5UTR (5'UTR) - TU2511
map/slr0786 (mRNA / CDS) - TU3208
slr0788 (mRNA / CDS)
slr0788-as (asRNA)
slr0789 (mRNA / CDS) - TU3212
slr0789-5UTR (5'UTR) - TU3212
umuC/slr0790 (mRNA / CDS) - TU3212
nrsB/slr0793 (mRNA / CDS)
nrsA/slr0794 (mRNA / CDS)
nrsD/slr0796 (mRNA / CDS) - TU3220
coaT/slr0797 (mRNA / CDS)
ziaA/slr0798 (mRNA / CDS)
slr0799 (mRNA / CDS) - TU3234
gcp/slr0807 (mRNA / CDS) - TU1728
slr0813 (mRNA / CDS) - TU1745
menF/slr0817 (mRNA / CDS) - TU1752
slr0820 (mRNA / CDS) - TU1757
alr/slr0827 (mRNA / CDS) - TU2986
slr0827-5UTR (5'UTR) - TU2986
slr0845 (mRNA / CDS) - TU3016
slr0845-5UTR (5'UTR) - TU3016
ndbA/slr0851 (mRNA / CDS) - TU1375
rimI/slr0853 (mRNA / CDS) - TU1377
psr/slr0883 (mRNA / CDS) - TU1184
slr0888 (mRNA / CDS) - TU1196
slr0888-5UTR (5'UTR) - TU1196
norM/slr0896 (mRNA / CDS) - TU2918
slr0897 (mRNA / CDS)
moaC/slr0902 (mRNA / CDS) - TU2925
bchE/slr0905 (mRNA / CDS) - TU2927
slr0921 (mRNA / CDS) - TU2941
ubiA/slr0926 (mRNA / CDS) - TU3400
slr0944 (mRNA / CDS) - TU2081
slr0950 (mRNA / CDS)
mrnC/slr0954 (mRNA / CDS) - TU2098
slr0957 (mRNA / CDS) - TU2100
slr0964 (mRNA / CDS) - TU320
slr0975 (mRNA / CDS) - TU342
slr0978 (mRNA / CDS) - TU350
slr0980 (mRNA / CDS) - TU352
slr1002 (mRNA / CDS)
slr1003 (mRNA / CDS)
slr1004 (mRNA / CDS)
slr1005 (mRNA / CDS)
slr1050 (mRNA / CDS) - TU489
slr1052 (mRNA / CDS) - TU490
slr1062 (mRNA / CDS) - TU837
slr1068 (mRNA / CDS) - TU355
slr1074 (mRNA / CDS) - TU355
galE/slr1078 (mRNA / CDS) - TU365
slr1082 (mRNA / CDS) - TU369
folK/slr1093 (mRNA / CDS) - TU181
slr1100 (mRNA / CDS) - TU188
slr1100-5UTR (5'UTR) - TU188
slr1119 (mRNA / CDS) - TU65
slr1119-5UTR (5'UTR) - TU65
hofD/slr1120 (mRNA / CDS) - TU71
slr1120-5UTR (5'UTR) - TU71
rhnB/slr1130 (mRNA / CDS) - TU87
slr1134 (mRNA / CDS) - TU786
slr1135 (mRNA / CDS) - TU788
ctaEI/slr1138 (mRNA / CDS) - TU791
gltS/slr1145 (mRNA / CDS)
slr1146 (mRNA / CDS)
hik2/slr1147 (mRNA / CDS) - TU810
slr1147-as2 (asRNA)
slr1152 (mRNA / CDS) - TU1982
slr1152-3UTR (3'UTR) - TU1982
slr1152-5UTR (5'UTR) - TU1982
slr1163 (mRNA / CDS) - TU2000
gldA/slr1167 (mRNA / CDS) - TU2009
slr1168 (mRNA / CDS) - TU2009
slr1170 (mRNA / CDS) - TU2010
slr1174 (mRNA / CDS) - TU2015
slr1174-5UTR (5'UTR) - TU2015
slr1182 (mRNA / CDS)
slr1187 (mRNA / CDS) - TU3686
slr1187-5UTR (5'UTR) - TU3686
mutL/slr1199 (mRNA / CDS) - TU886
livH/slr1200 (mRNA / CDS) - TU888
slr1200-5UTR (5'UTR) - TU888
slr1213 (mRNA / CDS)
slr1213-as (asRNA)
slr1214 (mRNA / CDS) - TU905
slr1215 (mRNA / CDS) - TU907
slr1215-5UTR (5'UTR) - TU907
slr1222 (mRNA / CDS) - TU1041
slr1229 (mRNA / CDS) - TU1060
slr1230 (mRNA / CDS) - TU1060
slr1243 (mRNA / CDS) - TU1417
slr1245 (mRNA / CDS)
slr1246 (mRNA / CDS) - TU1421
pstC/slr1248 (mRNA / CDS) - TU1428
pstA/slr1249 (mRNA / CDS) - TU1428
pstB/slr1250 (mRNA / CDS) - TU1428
slr1262 (mRNA / CDS) - TU1446
ftsW/slr1267 (mRNA / CDS) - TU1938
ycf62/slr1278 (mRNA / CDS)
ndhC/slr1279 (mRNA / CDS) - TU1954
slr1283 (mRNA / CDS) - TU1956
hik34/slr1285 (mRNA / CDS) - TU1957
ndhD2/slr1291 (mRNA / CDS) - TU282
slr1291-3UTR (3'UTR) - TU282
slr1291-5UTR (5'UTR) - TU282
slr1293 (mRNA / CDS) - TU285
slr1298 (mRNA / CDS)
slr1303 (mRNA / CDS) - TU303
slr1306 (mRNA / CDS) - TU309
slr1306-3UTR (3'UTR) - TU309
slr1306-5UTR (5'UTR) - TU309
fecC/slr1316 (mRNA / CDS) - TU19
fecD/slr1317 (mRNA / CDS) - TU21
fecB/slr1319 (mRNA / CDS) - TU23
fabF/slr1332 (mRNA / CDS) - TU1711
desA/slr1350 (mRNA / CDS) - TU1803
bioY/slr1365 (mRNA / CDS) - TU1139
lspA/slr1366 (mRNA / CDS) - TU1139
cdsA/slr1369 (mRNA / CDS) - TU1152
slr1369-5UTR (5'UTR) - TU1152
slr1376 (mRNA / CDS) - TU671
slr1376-5UTR (5'UTR) - TU671
cydB/slr1380 (mRNA / CDS) - TU672
feoB/slr1392 (mRNA / CDS) - TU689
slr1396 (mRNA / CDS) - TU698
slr1429 (mRNA / CDS) - TU3464
slr1436 (mRNA / CDS) - TU1090
slr1442 (mRNA / CDS) - TU1108
slr1444 (mRNA / CDS) - TU1109
slr1449 (mRNA / CDS) - TU1120
slr1451 (mRNA / CDS) - TU3471
cysA/slr1455 (mRNA / CDS)
slr1456 (mRNA / CDS) - TU3478
chrA/slr1457 (mRNA / CDS) - TU3480
slr1461 (mRNA / CDS) - TU3486
slr1461-5UTR (5'UTR) - TU3486
slr1484 (mRNA / CDS) - TU37
slr1485 (mRNA / CDS) - TU37
pchR/slr1489 (mRNA / CDS) - TU39
fhuA/slr1490 (mRNA / CDS) - TU42
slr1490-as2 (asRNA)
fecB/slr1491 (mRNA / CDS)
slr1491-as (asRNA)
slr1494 (mRNA / CDS)
slr1494-as (asRNA)
slr1495 (mRNA / CDS) - TU49
slr1495-5UTR (5'UTR) - TU49
slr1503 (mRNA / CDS) - TU1621
slr1508 (mRNA / CDS) - TU1634
ktrB, ntpJ/slr1509 (mRNA / CDS) - TU1634
menA/slr1518 (mRNA / CDS) - TU1651
slr1520 (mRNA / CDS) - TU1653
slr1522-5UTR (5'UTR) - TU1655
slr1530 (mRNA / CDS) - TU1969
slr1535 (mRNA / CDS)
recQ/slr1536 (mRNA / CDS) - TU991
cbiD/slr1538 (mRNA / CDS) - TU994
slr1543 (mRNA / CDS) - TU997
slr1544 (mRNA / CDS) - TU1000
slr1566 (mRNA / CDS) - TU3554
slr1567 (mRNA / CDS)
slr1583 (mRNA / CDS) - TU3578
slr1583-3UTR (3'UTR) - TU3578
slr1583-5UTR (5'UTR) - TU3578
slr1591 (mRNA / CDS)
slr1592 (mRNA / CDS) - TU431
slr1593 (mRNA / CDS) - TU435
slr1593-5UTR (5'UTR) - TU435
slr1594 (mRNA / CDS) - TU435
nhaS4/slr1595 (mRNA / CDS) - TU435
slr1601 (mRNA / CDS) - TU456
slr1601-3UTR (3'UTR) - TU456
slr1601-5UTR (5'UTR) - TU456
slr1627 (mRNA / CDS) - TU2119
slr1635-5UTR (5'UTR) - TU2129
slr1638 (mRNA / CDS) - TU2133
hlyB/slr1651 (mRNA / CDS)
slr1652 (mRNA / CDS) - TU3628
slr1674 (mRNA / CDS) - TU2024
slr1674-5UTR (5'UTR) - TU2024
slr1676 (mRNA / CDS) - TU2026
aspA/slr1705 (mRNA / CDS) - TU733
slr1705-5UTR (5'UTR) - TU733
slr1705-as (asRNA)
slr1708 (mRNA / CDS) - TU741
slr1726 (mRNA / CDS) - TU1520
slr1736 (mRNA / CDS) - TU1357
slr1737 (mRNA / CDS) - TU1357
slr1738 (mRNA / CDS) - TU1360
murI/slr1746 (mRNA / CDS) - TU1373
slr1746-5UTR (5'UTR) - TU1373
slr1747 (mRNA / CDS) - TU1374
hik14/slr1759 (mRNA / CDS)
slr1770 (mRNA / CDS) - TU528
slr1774 (mRNA / CDS)
slr1787 (mRNA / CDS)
pleD/slr1798 (mRNA / CDS) - TU257
mbpA/slr1803 (mRNA / CDS) - TU1308
slr1804 (mRNA / CDS) - TU1314
hik16/slr1805 (mRNA / CDS) - TU1314
slr1807 (mRNA / CDS) - TU1317
slr1807-5UTR (5'UTR) - TU1317
slr1815 (mRNA / CDS) - TU1328
nth/slr1822 (mRNA / CDS) - TU1344
slr1827 (mRNA / CDS) - TU936
slr1840 (mRNA / CDS) - TU969
merA/slr1849 (mRNA / CDS)
slr1851 (mRNA / CDS) - TU539
slr1851-5UTR (5'UTR) - TU539
slr1864 (mRNA / CDS) - TU1232
slr1871 (mRNA / CDS)
slr1885 (mRNA / CDS) - TU1276
ureF/slr1899 (mRNA / CDS) - TU858
slr1899-5UTR (5'UTR) - TU858
slr1906 (mRNA / CDS) - TU598
amiA/slr1910 (mRNA / CDS) - TU599
slr1914 (mRNA / CDS) - TU605
pilA7/slr1930 (mRNA / CDS) - TU2301
slr1935 (mRNA / CDS) - TU2317
dpm1/slr1943 (mRNA / CDS) - TU2336
ctaA/slr1950 (mRNA / CDS) - TU2348
slr1957 (mRNA / CDS) - TU1815
slr1966 (mRNA / CDS) - TU1831
slr1983 (mRNA / CDS) - TU1875
slr1990 (mRNA / CDS) - TU1476
slr1999 (mRNA / CDS) - TU1482
slr1999-5UTR (5'UTR) - TU1482
slr2003 (mRNA / CDS) - TU1488
slr2008 (mRNA / CDS) - TU757
slr2010 (mRNA / CDS) - TU757
slr2012 (mRNA / CDS) - TU757
slr2027 (mRNA / CDS) - TU776
slr2038 (mRNA / CDS)
slr2042 (mRNA / CDS) - TU562
slr2044 (mRNA / CDS)
slr2045 (mRNA / CDS)
phoH/slr2047 (mRNA / CDS) - TU580
apqZ/slr2057 (mRNA / CDS) - TU1454
slr2057-5UTR (5'UTR) - TU1454
slr2062-as (asRNA)
ctaEII/slr2083 (mRNA / CDS) - TU1573
slr2103 (mRNA / CDS) - TU1607
hik22/slr2104 (mRNA / CDS) - TU1608
slr2104-3UTR (3'UTR) - TU1608
kpsM/slr2107 (mRNA / CDS) - TU1665
spsC/slr2114 (mRNA / CDS) - TU1672
slr2115 (mRNA / CDS) - TU1673
slr2118 (mRNA / CDS) - TU1673
slr2126 (mRNA / CDS) - TU1677
hupE/slr2135 (mRNA / CDS) - TU1296
slr5010 (mRNA / CDS) - TU5022
slr5013 (mRNA / CDS)
slr5016 (mRNA / CDS) - TU5023
slr5029-3UTR (3'UTR) - TU5034
slr5038 (mRNA / CDS) - TU5039
slr5055 (mRNA / CDS) - TU5062
slr5056 (mRNA / CDS) - TU5062
slr5093 (mRNA / CDS)
slr5101 (mRNA / CDS) - TU5104
slr5112 (mRNA / CDS) - TU5117
slr5115 (mRNA / CDS) - TU5121
slr6001 (mRNA / CDS)
slr6013 (mRNA / CDS) - TU6009
slr6016 (mRNA / CDS) - TU6009
slr6021 (mRNA / CDS) - TU6020
slr6022 (mRNA / CDS) - TU6022
slr6031 (mRNA / CDS) - TU6031
slr6043 (mRNA / CDS)
slr6072 (mRNA / CDS) - TU6064
slr6075 (mRNA / CDS) - TU6064
slr6080 (mRNA / CDS) - TU6075
slr6081 (mRNA / CDS) - TU6077
slr6090 (mRNA / CDS) - TU6084
slr6106 (mRNA / CDS)
slr6108 (mRNA / CDS)
slr7005 (mRNA / CDS)
slr7010 (mRNA / CDS) - TU7004
slr7023 (mRNA / CDS) - TU7011
slr7025 (mRNA / CDS) - TU7011
slr7037 (mRNA / CDS) - TU7032
slr7049 (mRNA / CDS) - TU7044
slr7052 (mRNA / CDS)
slr7054 (mRNA / CDS)
slr7057 (mRNA / CDS) - TU7053
slr7057-5UTR (5'UTR) - TU7053
slr7058 (mRNA / CDS) - TU7053
slr7059 (mRNA / CDS) - TU7054
slr7068 (mRNA / CDS) - TU7064
cas1/slr7071 (mRNA / CDS) - TU7066
slr7076 (mRNA / CDS)
slr7081 (mRNA / CDS) - TU7075
slr7082 (mRNA / CDS) - TU7075
slr7083 (mRNA / CDS)
cas1/slr7092 (mRNA / CDS) - TU7084
slr7097 (mRNA / CDS) - TU7089
slr7100 (mRNA / CDS) - TU7091
slr7101 (mRNA / CDS)
slr8016 (mRNA / CDS) - TU8009
slr8021 (mRNA / CDS) - TU8022
slr8026 (mRNA / CDS) - TU8023
slr8029 (mRNA / CDS) - TU8025
slr8036 (mRNA / CDS) - TU8030
slr8037 (mRNA / CDS) - TU8032
slr8038 (mRNA / CDS) - TU8034
slr8044 (mRNA / CDS)
psbI/sml0001 (mRNA / CDS) - TU2443
sml0001-3UTR (3'UTR) - TU2443
sml0001-5UTR (5'UTR) - TU2443
psbX/sml0002 (mRNA / CDS) - TU2754
sml0002-5UTR (5'UTR) - TU2754
psbM/sml0003 (mRNA / CDS) - TU134
sml0003-3UTR (3'UTR) - TU134
sml0003-5UTR (5'UTR) - TU134
petN/sml0004 (mRNA / CDS) - TU149
psbK/sml0005 (mRNA / CDS) - TU530
sml0012 (mRNA / CDS) - TU908
psbT, ycf8/smr0001 (mRNA / CDS) - TU2512
smr0001-3UTR (3'UTR) - TU2512
smr0001-5UTR (5'UTR) - TU2512
psaI/smr0004 (mRNA / CDS) - TU3633
psaM/smr0005 (mRNA / CDS) - TU455
smr0005-3UTR (3'UTR) - TU455
smr0005-5UTR (5'UTR) - TU455
psbJ/smr0008 (mRNA / CDS) - TU548
psbN/smr0009 (mRNA / CDS) - TU1204
smr0009-3UTR (3'UTR) - TU1204
smr0009-5UTR (5'UTR) - TU1204
petG/smr0010 (mRNA / CDS) - TU1891
smr0010-5UTR (5'UTR) - TU1891
rpl34/smr0011 (mRNA / CDS) - TU1898
smr0011-5UTR (5'UTR) - TU1898
smr0015 (mRNA / CDS) - TU2809
ssl0296 (mRNA / CDS)
ssl0318 (mRNA / CDS)
ssl0353 (mRNA / CDS) - TU2892
ssl0353-3UTR (3'UTR) - TU2892
nblA1/ssl0452 (mRNA / CDS) - TU1562
ssl0452-5UTR (5'UTR) - TU1562
nblA2/ssl0453 (mRNA / CDS) - TU1562
ssl0606 (mRNA / CDS)
ssl0769-3UTR (3'UTR) - TU2686
ssl0900 (mRNA / CDS) - TU2744
ssl1047 (mRNA / CDS) - TU2959
ssl1047-5UTR (5'UTR) - TU2959
ssl1300 (mRNA / CDS) - TU2778
ycf33/ssl1417 (mRNA / CDS) - TU2514
ssl1493 (mRNA / CDS)
ssl1577-5UTR (5'UTR) - TU1407
ssl1807 (mRNA / CDS) - TU2358
ssl1807-3UTR (3'UTR) - TU2358
ssl1807-5UTR (5'UTR) - TU2358
gifA/ssl1911 (mRNA / CDS) - TU627
ssl1911-3UTR (3'UTR) - TU627
ssl1911-5UTR (5'UTR) - TU627
ssl2100 (mRNA / CDS) - TU805
ssl2162 (mRNA / CDS) - TU1983
ssl2162-3UTR (3'UTR) - TU1983
ssl2162-5UTR (5'UTR) - TU1983
ssl2250 (mRNA / CDS) - TU1071
ssl2250-3UTR (3'UTR) - TU1071
ssl2380 (mRNA / CDS) - TU291
ssl2384 (mRNA / CDS) - TU289
ssl2502 (mRNA / CDS) - TU1144
hliA, scpC/ssl2542 (mRNA / CDS) - TU690
ssl2559 (mRNA / CDS) - TU679
ssl2996-5UTR (5'UTR) - TU3663
ssl3076 (mRNA / CDS)
repA/ssl3177 (mRNA / CDS) - TU1030
ssl3177-3UTR (3'UTR) - TU1030
ssl3177-5UTR (5'UTR) - TU1030
ssl3382 (mRNA / CDS) - TU1249
ssl3383 (mRNA / CDS) - TU1249
ssl3410 (mRNA / CDS) - TU1224
ssl3446 (mRNA / CDS) - TU830
ssl3573 (mRNA / CDS) - TU1823
ssl3573-3UTR (3'UTR) - TU1823
hypC/ssl3580 (mRNA / CDS) - TU1817
ssl3580-5UTR (5'UTR) - TU1817
ssl3769 (mRNA / CDS) - TU1593
ssl3803 (mRNA / CDS) - TU1681
ssl5027 (mRNA / CDS) - TU5031
ssl5098 (mRNA / CDS) - TU5102
ssl5099 (mRNA / CDS) - TU5103
ssl5114 (mRNA / CDS) - TU5119
ssl5129 (mRNA / CDS) - TU5130
ssl6092 (mRNA / CDS) - TU6086
ssl7074 (mRNA / CDS) - TU7071
ssl8010 (mRNA / CDS) - TU8008
ssr0336 (mRNA / CDS) - TU2913
ssr0511 (mRNA / CDS)
ssr0536 (mRNA / CDS) - TU2380
ssr0663-5UTR (5'UTR) - TU2240
ssr0755 (mRNA / CDS) - TU2162
ssr0759 (mRNA / CDS) - TU2163
ssr1038 (mRNA / CDS) - TU3113
ssr1038-3UTR (3'UTR) - TU3113
ssr1038-5UTR (5'UTR) - TU3113
ssr1155 (mRNA / CDS) - TU422
ESI3/ssr1169 (mRNA / CDS) - TU3254
ssr1169-5UTR (5'UTR) - TU3254
ssr1375 (mRNA / CDS) - TU1723
ssr1375-3UTR (3'UTR) - TU1723
ssr1407 (mRNA / CDS) - TU2993
ssr1407-5UTR (5'UTR) - TU2993
ssr1562 (mRNA / CDS) - TU3408
ssr1562-3UTR (3'UTR) - TU3408
ssr1562-5UTR (5'UTR) - TU3408
ssr1880 (mRNA / CDS) - TU786
ssr1966 (mRNA / CDS)
ssr2078 (mRNA / CDS) - TU1421
ssr2078-5UTR (5'UTR) - TU1421
ssr2130 (mRNA / CDS) - TU1960
ycf19/ssr2142 (mRNA / CDS) - TU286
ssr2142-3UTR (3'UTR) - TU286
ssr2153 (mRNA / CDS) - TU300
ssr2153-3UTR (3'UTR) - TU300
ssr2153-5UTR (5'UTR) - TU300
ssr2194 (mRNA / CDS) - TU1693
ssr2194-3UTR (3'UTR) - TU1693
ssr2315 (mRNA / CDS) - TU674
ssr2333 (mRNA / CDS) - TU689
ssr2340 (mRNA / CDS) - TU695
ssr2549-3UTR (3'UTR) - TU1644
ssr2553 (mRNA / CDS) - TU1649
hliB, scpD/ssr2595 (mRNA / CDS) - TU1000
ssr2595-5UTR (5'UTR) - TU1000
ssr2611 (mRNA / CDS) - TU1015
ssr2611-5UTR (5'UTR) - TU1015
ssr2615 (mRNA / CDS)
ssr2708 (mRNA / CDS) - TU469
ssr2710 (mRNA / CDS) - TU469
ssr2784-3UTR (3'UTR) - TU3656
ssr2786 (mRNA / CDS) - TU3661
ssr2806-5UTR (5'UTR) - TU2032
ssr2843 (mRNA / CDS) - TU710
ssr2848 (mRNA / CDS) - TU724
ssr2848-3UTR (3'UTR) - TU724
ssr2899 (mRNA / CDS) - TU1516
ssr2912 (mRNA / CDS) - TU1526
ssr3129 (mRNA / CDS) - TU539
ssr3184 (mRNA / CDS) - TU1269
secG, ycf47/ssr3307 (mRNA / CDS) - TU2341
ssr3307-3UTR (3'UTR) - TU2341
ssr3409 (mRNA / CDS) - TU757
ssr3410 (mRNA / CDS) - TU757
ssr3452 (mRNA / CDS)
ssr3465 (mRNA / CDS) - TU567
ssr3465-5UTR (5'UTR) - TU567
ssr3571 (mRNA / CDS) - TU1665
ssr3572 (mRNA / CDS) - TU1665
ssr5117 (mRNA / CDS) - TU5121
ssr5120 (mRNA / CDS) - TU5122
ssr6020 (mRNA / CDS)
ssr6030 (mRNA / CDS) - TU6027
ssr6030-3UTR (3'UTR) - TU6027
ssr6079 (mRNA / CDS)
ssr6089-3UTR (3'UTR) - TU6082
ssr6099 (mRNA / CDS) - TU6093
ssr7036 (mRNA / CDS) - TU7030
ssr7040 (mRNA / CDS) - TU7034
ssr8013-5UTR (5'UTR) - TU8009